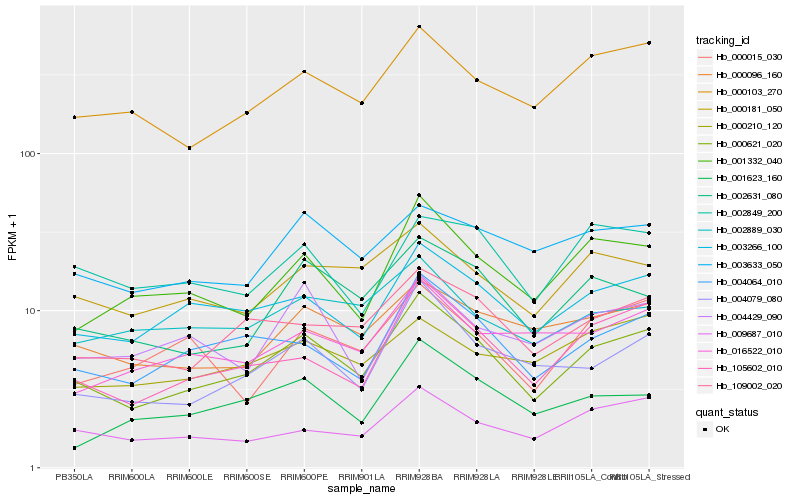
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000621_020 |
0.0 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 2 |
Hb_003266_100 |
0.1018229834 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 3 |
Hb_004064_010 |
0.1028977567 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 4 |
Hb_105602_010 |
0.1033374865 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 5 |
Hb_109002_020 |
0.1074261939 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 6 |
Hb_004079_080 |
0.108630105 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 7 |
Hb_000103_270 |
0.1119863398 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_002631_080 |
0.1123372433 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_001332_040 |
0.1156176253 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000096_160 |
0.1215128547 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 11 |
Hb_000181_050 |
0.1226544279 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 12 |
Hb_009687_010 |
0.123562809 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_003633_050 |
0.1289876901 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 14 |
Hb_001623_160 |
0.1322788896 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 15 |
Hb_016522_010 |
0.1362691677 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 16 |
Hb_002889_030 |
0.1364611739 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 17 |
Hb_002849_200 |
0.1366892848 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 18 |
Hb_000210_120 |
0.1386065714 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004429_090 |
0.1390988398 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000015_030 |
0.1394721995 |
- |
- |
PREDICTED: THO complex subunit 4B-like [Jatropha curcas] |