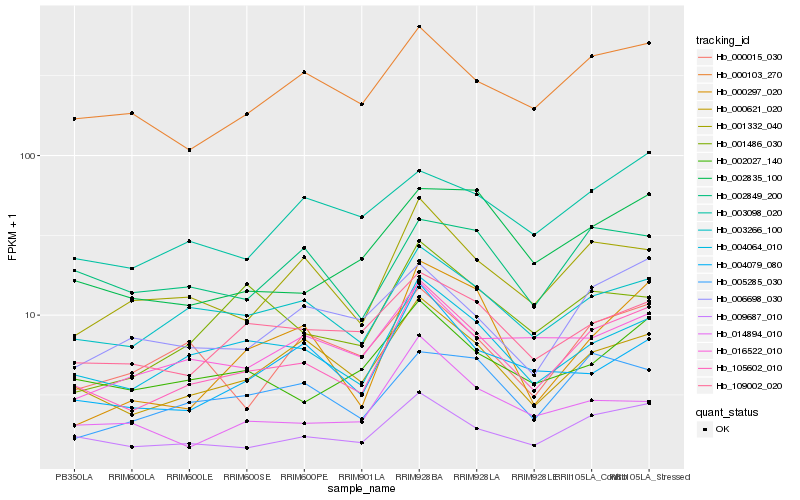
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105602_010 |
0.0 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 2 |
Hb_009687_010 |
0.0997602878 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 3 |
Hb_004064_010 |
0.1009359678 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 4 |
Hb_000621_020 |
0.1033374865 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 5 |
Hb_003266_100 |
0.1173917535 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 6 |
Hb_001332_040 |
0.1205057961 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 7 |
Hb_000103_270 |
0.1294123646 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_002027_140 |
0.1309136392 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 9 |
Hb_109002_020 |
0.1365636391 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 10 |
Hb_004079_080 |
0.1376391346 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 11 |
Hb_006698_030 |
0.1441817649 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 12 |
Hb_002835_100 |
0.1467226708 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 13 |
Hb_016522_010 |
0.1479820927 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 14 |
Hb_001486_030 |
0.148281118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005285_030 |
0.1505962295 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 16 |
Hb_014894_010 |
0.1510337725 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 17 |
Hb_000015_030 |
0.1514646896 |
- |
- |
PREDICTED: THO complex subunit 4B-like [Jatropha curcas] |
| 18 |
Hb_003098_020 |
0.1518215509 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |
| 19 |
Hb_000297_020 |
0.1539927905 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 20 |
Hb_002849_200 |
0.1556770786 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |