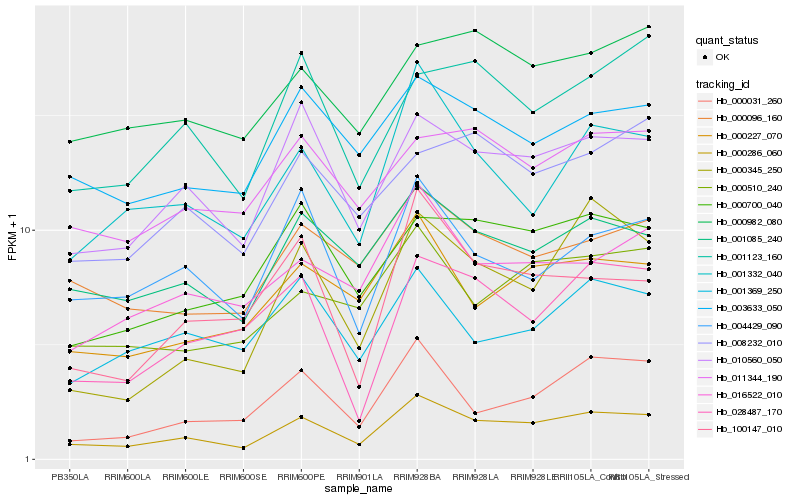
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000286_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 2 |
Hb_010560_050 |
0.0942643365 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 3 |
Hb_001085_240 |
0.1000845231 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001332_040 |
0.1081764163 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 5 |
Hb_000227_070 |
0.1112077004 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 6 |
Hb_003633_050 |
0.1152780653 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 7 |
Hb_000096_160 |
0.1156294112 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 8 |
Hb_016522_010 |
0.1156804494 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 9 |
Hb_000510_240 |
0.1157360354 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_004429_090 |
0.1168109588 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000700_040 |
0.1172425688 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 12 |
Hb_000031_260 |
0.1206020703 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 13 |
Hb_011344_190 |
0.1241913735 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 14 |
Hb_100147_010 |
0.1254972436 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 15 |
Hb_028487_170 |
0.1262719072 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 16 |
Hb_001123_160 |
0.1275634154 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 17 |
Hb_001369_250 |
0.1278660295 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000345_250 |
0.1291467777 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 19 |
Hb_000982_080 |
0.1295392857 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 20 |
Hb_008232_010 |
0.1304400098 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |