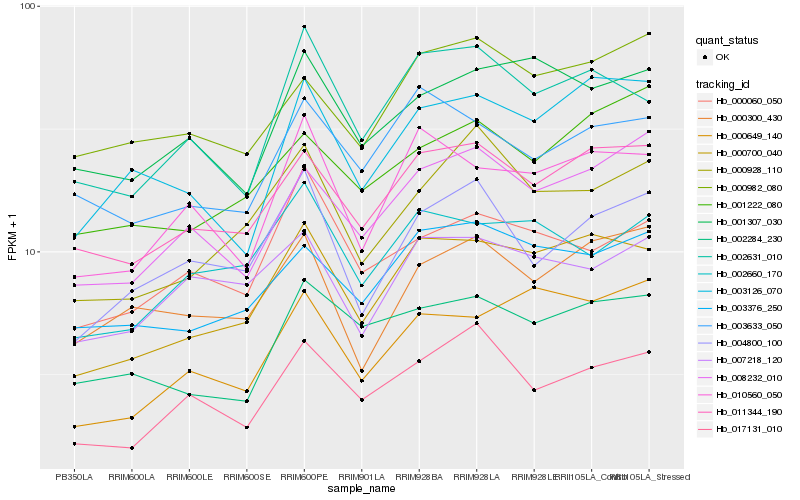
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_040 |
0.0 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |
| 2 |
Hb_011344_190 |
0.0754529737 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 3 |
Hb_010560_050 |
0.0830097093 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 4 |
Hb_000649_140 |
0.0870640938 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002631_010 |
0.0871066492 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 6 |
Hb_003376_250 |
0.0875603297 |
- |
- |
Transducin/WD40 repeat-like superfamily protein [Theobroma cacao] |
| 7 |
Hb_003126_070 |
0.0938394845 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002660_170 |
0.0968280642 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_001307_030 |
0.0997054226 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000928_110 |
0.1000416066 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008232_010 |
0.1000955439 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 12 |
Hb_002284_230 |
0.100331452 |
- |
- |
PREDICTED: uncharacterized protein At5g43822 [Jatropha curcas] |
| 13 |
Hb_003633_050 |
0.1010913285 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 14 |
Hb_000982_080 |
0.1077567906 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 15 |
Hb_017131_010 |
0.110260613 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 16 |
Hb_007218_120 |
0.1106134321 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000060_050 |
0.1115665085 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 18 |
Hb_001222_080 |
0.1117687591 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000300_430 |
0.1119821165 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 20 |
Hb_004800_100 |
0.1125948258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |