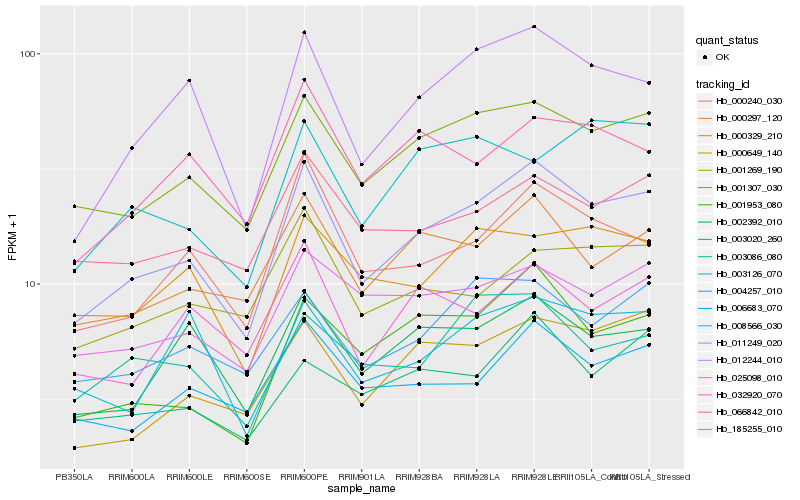
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011249_020 |
0.0 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 2 |
Hb_001307_030 |
0.0767313463 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 3 |
Hb_012244_010 |
0.0831250665 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 4 |
Hb_000297_120 |
0.0966403857 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 5 |
Hb_000240_030 |
0.0978576753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000649_140 |
0.0986915176 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006683_070 |
0.1012344912 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 8 |
Hb_001953_080 |
0.1015874559 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 9 |
Hb_008566_030 |
0.1017366212 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 10 |
Hb_002392_010 |
0.1017887793 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 11 |
Hb_185255_010 |
0.1060298116 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 12 |
Hb_025098_010 |
0.1069521613 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000329_210 |
0.1103981158 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 14 |
Hb_003020_260 |
0.1110788467 |
- |
- |
PREDICTED: metal tolerance protein B [Jatropha curcas] |
| 15 |
Hb_032920_070 |
0.1117324354 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 16 |
Hb_003086_080 |
0.1142475366 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 17 |
Hb_001269_190 |
0.1153644127 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 18 |
Hb_066842_010 |
0.1159659092 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004257_010 |
0.1162123141 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 20 |
Hb_003126_070 |
0.1182030248 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |