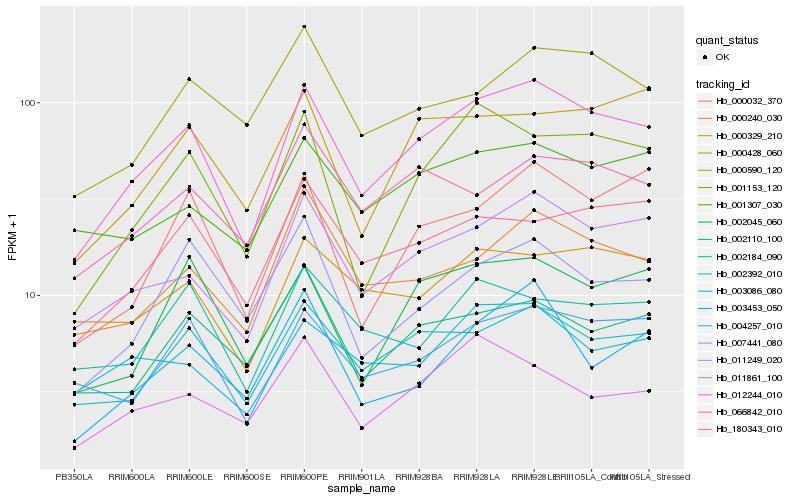
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012244_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 2 |
Hb_011249_020 |
0.0831250665 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 3 |
Hb_002392_010 |
0.0909545276 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 4 |
Hb_004257_010 |
0.1036720538 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 5 |
Hb_001153_120 |
0.10613579 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 6 |
Hb_180343_010 |
0.1120236856 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 7 |
Hb_000240_030 |
0.1142686594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002045_060 |
0.1152411889 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001307_030 |
0.1169606548 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000032_370 |
0.1187730903 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 11 |
Hb_000329_210 |
0.1195880893 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 12 |
Hb_002110_100 |
0.1199442177 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000428_060 |
0.1214801399 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000590_120 |
0.1223322967 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 15 |
Hb_003086_080 |
0.1227974154 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 16 |
Hb_066842_010 |
0.123903727 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003453_050 |
0.1250499308 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 18 |
Hb_002184_090 |
0.1255431055 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_011861_100 |
0.1259503761 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_007441_080 |
0.1260157218 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |