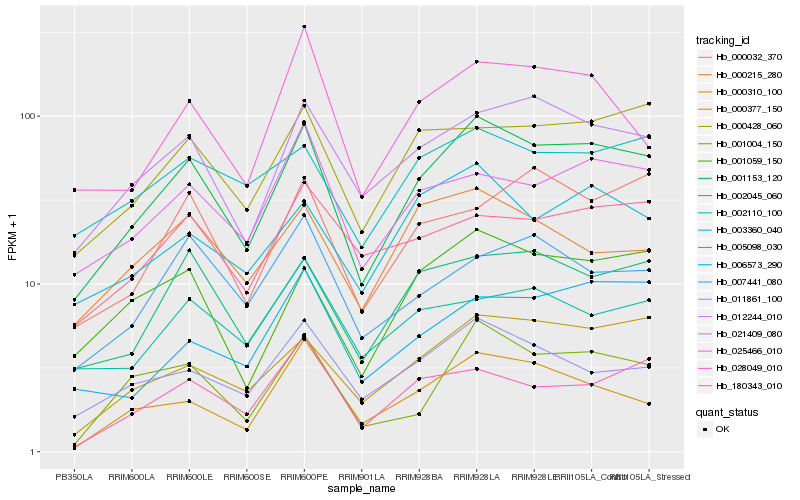
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_120 |
0.0 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 2 |
Hb_012244_010 |
0.10613579 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 3 |
Hb_001059_150 |
0.1159762103 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 4 |
Hb_002045_060 |
0.1168846457 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000377_150 |
0.1169884762 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 6 |
Hb_000428_060 |
0.1180636347 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_011861_100 |
0.1250455851 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_021409_080 |
0.126136972 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 9 |
Hb_001004_150 |
0.1272001209 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 10 |
Hb_005098_030 |
0.1279613881 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003360_040 |
0.1374950543 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 12 |
Hb_006573_290 |
0.1386689805 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 13 |
Hb_000310_100 |
0.1398924558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_025466_010 |
0.1399878214 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000032_370 |
0.1404749954 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 16 |
Hb_180343_010 |
0.1411486319 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 17 |
Hb_028049_010 |
0.1423869599 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007441_080 |
0.1451843978 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 19 |
Hb_000215_280 |
0.1461775221 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 20 |
Hb_002110_100 |
0.1479728494 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |