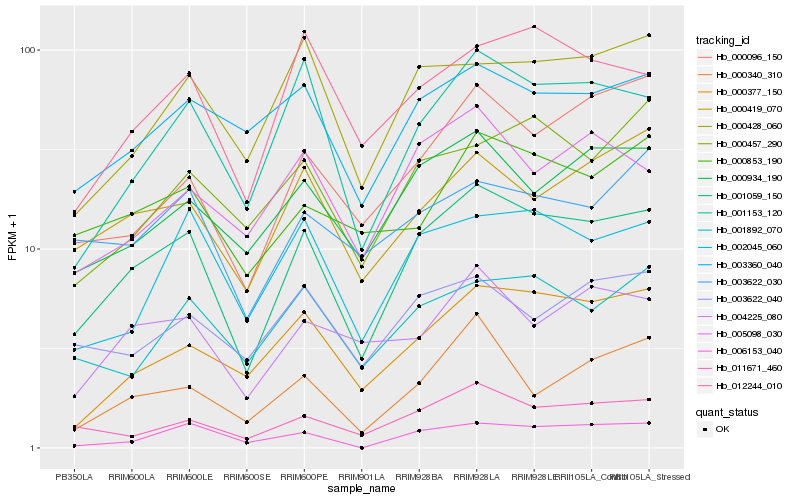
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001059_150 |
0.0 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 2 |
Hb_001153_120 |
0.1159762103 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 3 |
Hb_011671_460 |
0.1248409102 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 4 |
Hb_000934_190 |
0.1275097869 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 5 |
Hb_002045_060 |
0.1307753691 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000377_150 |
0.1316766248 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 7 |
Hb_000419_070 |
0.1323319123 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 8 |
Hb_012244_010 |
0.1341591908 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 9 |
Hb_000428_060 |
0.1363000435 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_003360_040 |
0.1377633755 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 11 |
Hb_001892_070 |
0.1402495653 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 12 |
Hb_005098_030 |
0.1404850537 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004225_080 |
0.142972118 |
- |
- |
- |
| 14 |
Hb_000853_190 |
0.146237331 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 15 |
Hb_003622_040 |
0.1479439309 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 16 |
Hb_000096_150 |
0.1485795219 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_006153_040 |
0.149172192 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |
| 18 |
Hb_000457_290 |
0.1509472091 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 19 |
Hb_003622_030 |
0.1516528556 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 1 [Jatropha curcas] |
| 20 |
Hb_000340_310 |
0.1518516371 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |