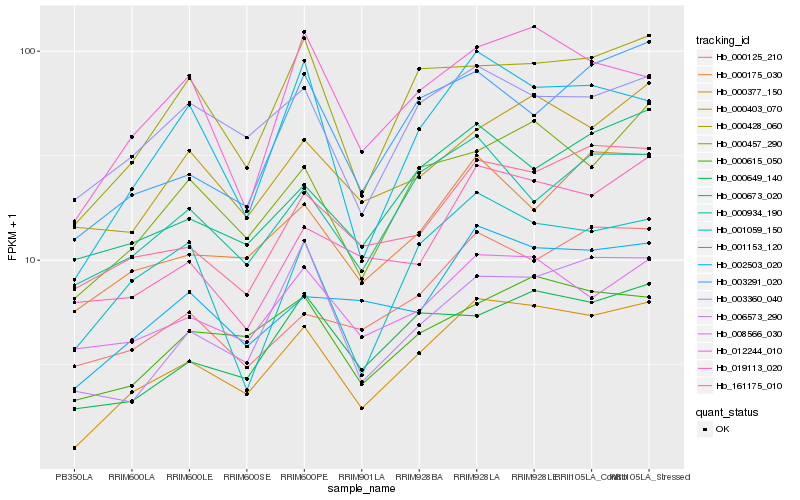
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000377_150 |
0.0 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 2 |
Hb_000457_290 |
0.1058007925 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 3 |
Hb_002503_020 |
0.1154790961 |
- |
- |
Phosphatase 2C family protein, putative [Theobroma cacao] |
| 4 |
Hb_000428_060 |
0.1157006308 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000615_050 |
0.1165302785 |
- |
- |
Potassium transporter 11 family protein [Populus trichocarpa] |
| 6 |
Hb_001153_120 |
0.1169884762 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 7 |
Hb_161175_010 |
0.1183497773 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 8 |
Hb_000649_140 |
0.1194600455 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000175_030 |
0.1205285758 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 10 |
Hb_000673_020 |
0.1270612771 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 11 |
Hb_003291_020 |
0.1281554711 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 12 |
Hb_000403_070 |
0.1289881365 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_019113_020 |
0.1293648326 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 14 |
Hb_000934_190 |
0.130746059 |
- |
- |
SER/ARG-rich protein 34A [Theobroma cacao] |
| 15 |
Hb_008566_030 |
0.131408628 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 16 |
Hb_001059_150 |
0.1316766248 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 17 |
Hb_012244_010 |
0.1320219163 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 18 |
Hb_000125_210 |
0.1327697308 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_003360_040 |
0.1329627258 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 20 |
Hb_006573_290 |
0.1334037574 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |