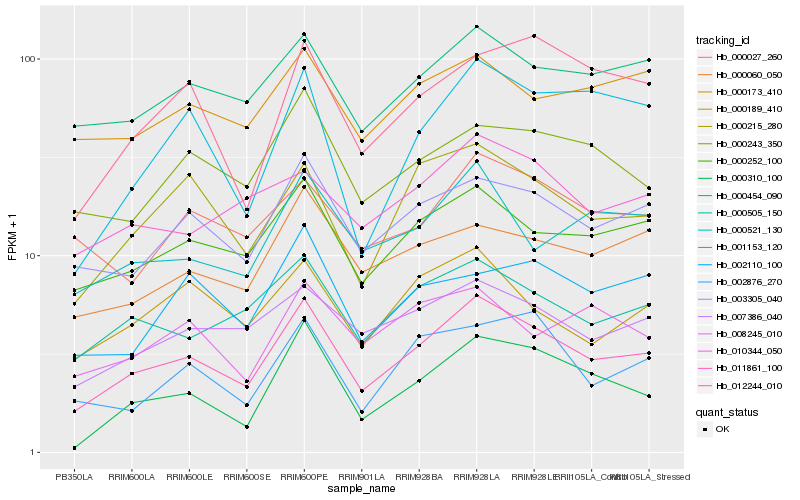
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011861_100 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000252_100 |
0.0953867953 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 3 |
Hb_003305_040 |
0.0983212757 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 4 |
Hb_000215_280 |
0.1047531557 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 5 |
Hb_000505_150 |
0.1064429164 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 6 |
Hb_000454_090 |
0.1089685113 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 7 |
Hb_007386_040 |
0.1111646104 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000060_050 |
0.1188219339 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 9 |
Hb_000521_130 |
0.1192556147 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000189_410 |
0.1196139694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000243_350 |
0.1199677069 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 12 |
Hb_000310_100 |
0.1230231476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001153_120 |
0.1250455851 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 14 |
Hb_012244_010 |
0.1259503761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 15 |
Hb_010344_050 |
0.1272101257 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 16 |
Hb_002110_100 |
0.1280363501 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002876_270 |
0.1289121386 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000173_410 |
0.1295144615 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 19 |
Hb_000027_260 |
0.1312173255 |
- |
- |
PREDICTED: chaperone protein dnaJ 15 [Jatropha curcas] |
| 20 |
Hb_008245_010 |
0.1323036004 |
- |
- |
PREDICTED: protein NEDD1 [Jatropha curcas] |