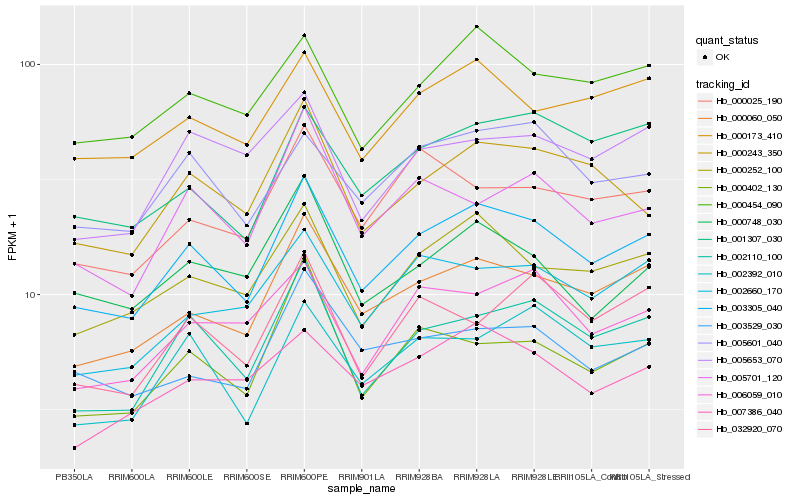
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003305_040 |
0.0 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 2 |
Hb_000060_050 |
0.0601509201 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 3 |
Hb_000252_100 |
0.067262291 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 4 |
Hb_002110_100 |
0.070061939 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000454_090 |
0.0761762638 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 6 |
Hb_000173_410 |
0.0789986864 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 7 |
Hb_005601_040 |
0.0804240679 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 8 |
Hb_000243_350 |
0.0828734274 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 9 |
Hb_002660_170 |
0.0876066912 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_001307_030 |
0.087873057 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002392_010 |
0.0890313584 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 12 |
Hb_006059_010 |
0.0895134202 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 13 |
Hb_003529_030 |
0.0906745214 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 14 |
Hb_000025_190 |
0.0918298026 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 15 |
Hb_007386_040 |
0.0920099262 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_005701_120 |
0.0925634672 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 17 |
Hb_000748_030 |
0.0937200364 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000402_130 |
0.0944824347 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 19 |
Hb_032920_070 |
0.094863877 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 20 |
Hb_005653_070 |
0.0952279079 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |