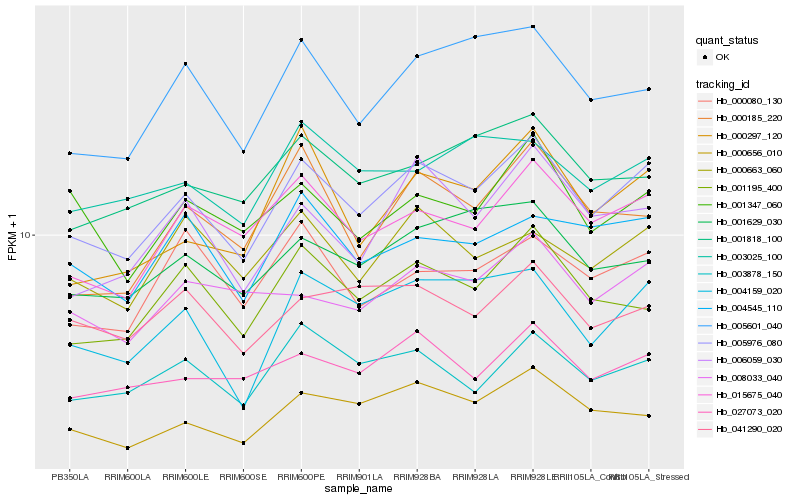
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005976_080 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 2 |
Hb_015675_040 |
0.0672462509 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 3 |
Hb_004159_020 |
0.0703304414 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 4 |
Hb_000656_010 |
0.0708771734 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003878_150 |
0.0742840557 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 6 |
Hb_027073_020 |
0.0758922005 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_001195_400 |
0.0773575362 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 8 |
Hb_005601_040 |
0.0778280381 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_001629_030 |
0.0795083017 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 10 |
Hb_006059_030 |
0.0800649887 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000663_060 |
0.0816991043 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 12 |
Hb_004545_110 |
0.0831340009 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_008033_040 |
0.08313592 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001818_100 |
0.0833270726 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 15 |
Hb_041290_020 |
0.0855904171 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_001347_060 |
0.0862983866 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 17 |
Hb_000080_130 |
0.0870203628 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 18 |
Hb_003025_100 |
0.087292218 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000297_120 |
0.0874772632 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 20 |
Hb_000185_220 |
0.0880813466 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |