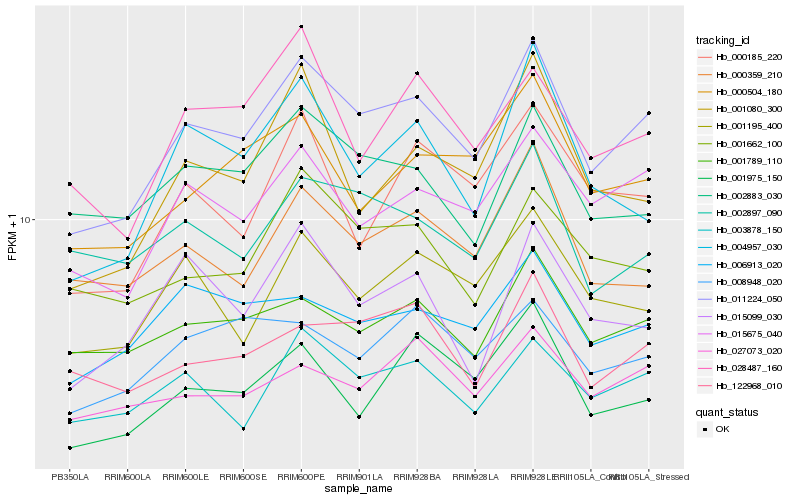
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011224_050 |
0.0 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 2 |
Hb_015675_040 |
0.0678114083 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 3 |
Hb_003878_150 |
0.0784934108 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 4 |
Hb_001195_400 |
0.0832764894 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 5 |
Hb_001975_150 |
0.0833796997 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 6 |
Hb_000504_180 |
0.0852606855 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 7 |
Hb_028487_160 |
0.0853280703 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 8 |
Hb_001789_110 |
0.0854079438 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_001662_100 |
0.085486563 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 10 |
Hb_122968_010 |
0.0857069189 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 11 |
Hb_006913_020 |
0.0863591594 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002883_030 |
0.0877876679 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 13 |
Hb_000359_210 |
0.0884405984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008948_020 |
0.0885913125 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 15 |
Hb_001080_300 |
0.0891750075 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 16 |
Hb_015099_030 |
0.0898008391 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 17 |
Hb_000185_220 |
0.0898329371 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 18 |
Hb_004957_030 |
0.0902627361 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 19 |
Hb_027073_020 |
0.0906681155 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_002897_090 |
0.0921099644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |