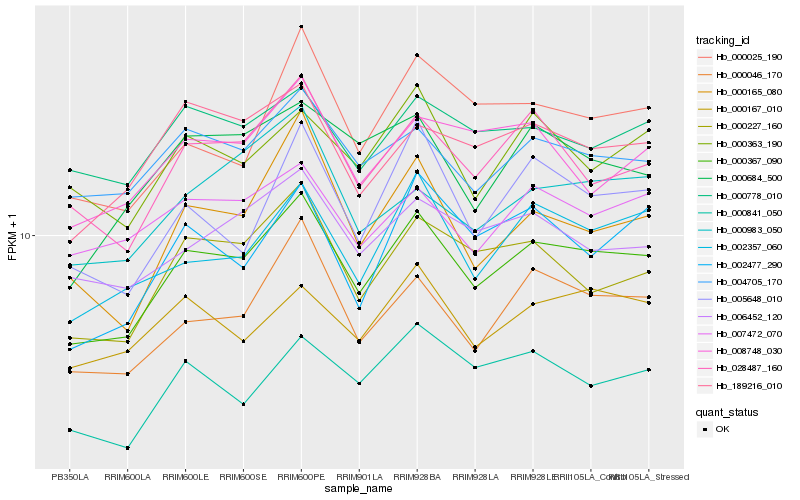
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_090 |
0.0 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 2 |
Hb_000046_170 |
0.068259911 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 3 |
Hb_028487_160 |
0.0691796525 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 4 |
Hb_000227_160 |
0.0717163135 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 5 |
Hb_002357_060 |
0.0721193123 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 6 |
Hb_004705_170 |
0.0756928224 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 7 |
Hb_189216_010 |
0.079124756 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000165_080 |
0.0793381146 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006452_120 |
0.0806869671 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 10 |
Hb_000167_010 |
0.0811483512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005648_010 |
0.0821597823 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 12 |
Hb_007472_070 |
0.0832137208 |
- |
- |
cir, putative [Ricinus communis] |
| 13 |
Hb_000684_500 |
0.0840494094 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_002477_290 |
0.0843081703 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000841_050 |
0.0846187028 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 16 |
Hb_000363_190 |
0.0847721775 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_008748_030 |
0.0849801568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000778_010 |
0.0853430782 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 19 |
Hb_000025_190 |
0.0855190369 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_000983_050 |
0.0861455745 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |