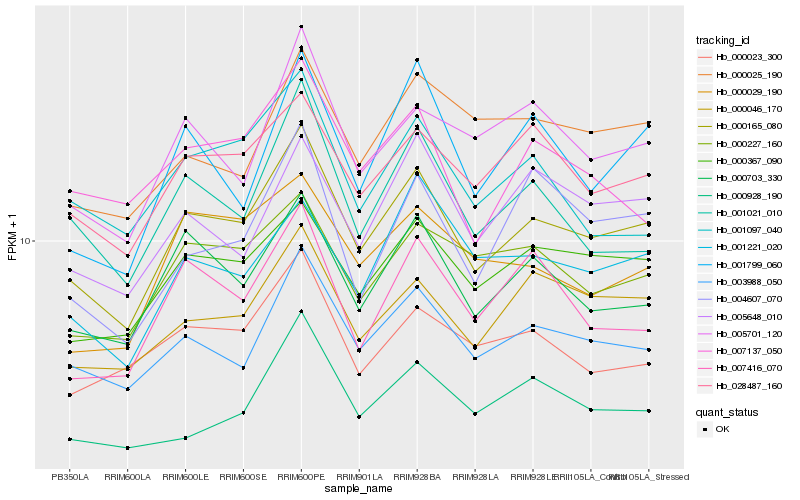
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_080 |
0.0 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003988_050 |
0.0697174844 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 3 |
Hb_000367_090 |
0.0793381146 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 4 |
Hb_000703_330 |
0.0797507579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000046_170 |
0.0825636966 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 6 |
Hb_028487_160 |
0.083266154 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 7 |
Hb_000227_160 |
0.0923291466 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 8 |
Hb_005701_120 |
0.0927040559 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 9 |
Hb_001021_010 |
0.0964003927 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 10 |
Hb_005648_010 |
0.0971685545 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_300 |
0.0979921778 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 12 |
Hb_007416_070 |
0.1010313893 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 13 |
Hb_000025_190 |
0.1014007576 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 14 |
Hb_001221_020 |
0.1014574839 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 15 |
Hb_007137_050 |
0.1021790961 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_001799_060 |
0.1026438405 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 17 |
Hb_000029_190 |
0.1038154599 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 18 |
Hb_004607_070 |
0.1051626764 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 19 |
Hb_001097_040 |
0.1051703944 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 20 |
Hb_000928_190 |
0.1061885105 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |