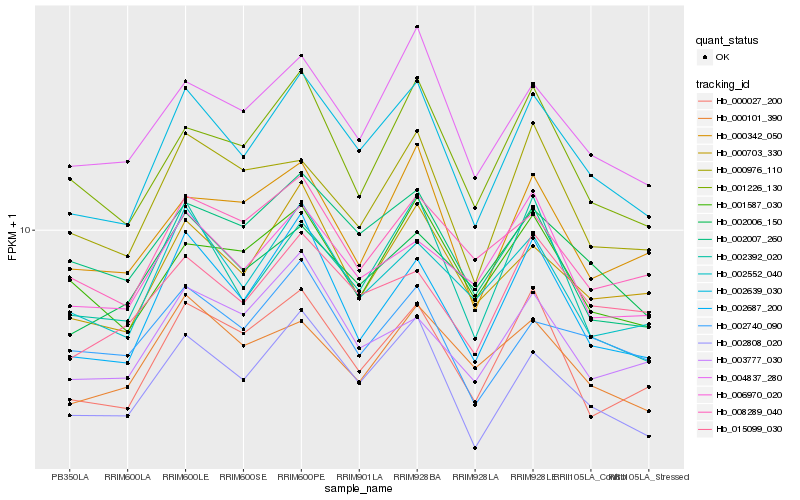
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_030 |
0.0 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 2 |
Hb_002808_020 |
0.0725883422 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004837_280 |
0.0782667199 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 4 |
Hb_000703_330 |
0.078671286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002687_200 |
0.0793781857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001226_130 |
0.0859180805 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_002552_040 |
0.0876542329 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 8 |
Hb_015099_030 |
0.0902214941 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 9 |
Hb_000027_200 |
0.0913915902 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 10 |
Hb_002392_020 |
0.0931738925 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 11 |
Hb_000976_110 |
0.093522767 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 12 |
Hb_002007_260 |
0.0946694177 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 13 |
Hb_001587_030 |
0.0956959097 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 14 |
Hb_000342_050 |
0.0959942542 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 15 |
Hb_002006_150 |
0.0965913959 |
- |
- |
copine, putative [Ricinus communis] |
| 16 |
Hb_008289_040 |
0.0971360303 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 17 |
Hb_003777_030 |
0.0971846302 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 18 |
Hb_002740_090 |
0.0972164772 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 19 |
Hb_000101_390 |
0.0975589973 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 20 |
Hb_006970_020 |
0.0988118502 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |