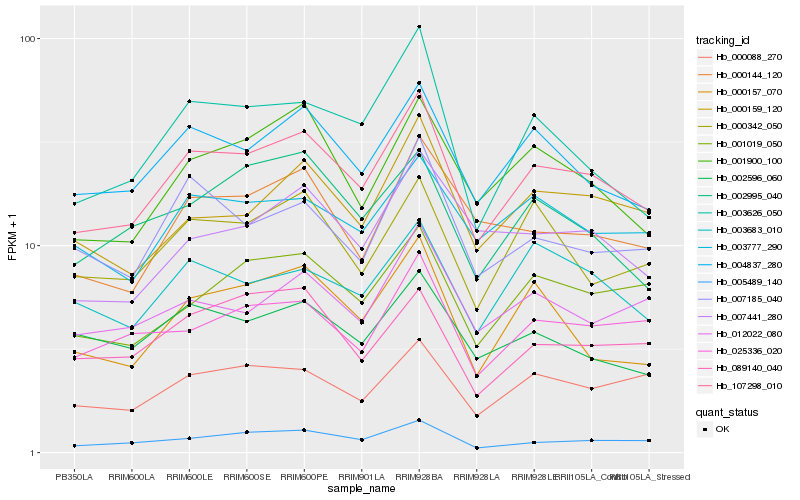
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107298_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 2 |
Hb_004837_280 |
0.073622656 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 3 |
Hb_005489_140 |
0.0816693494 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 4 |
Hb_001900_100 |
0.0912328224 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000159_120 |
0.0928541357 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 6 |
Hb_001019_050 |
0.093116419 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 7 |
Hb_003777_290 |
0.0949167119 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 8 |
Hb_000088_270 |
0.0956605977 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 9 |
Hb_025336_020 |
0.0957331829 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 10 |
Hb_000342_050 |
0.0979151144 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 11 |
Hb_003626_050 |
0.0985647659 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 12 |
Hb_007185_040 |
0.0995452831 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 13 |
Hb_003683_010 |
0.0997778942 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 14 |
Hb_000144_120 |
0.1008120886 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_012022_080 |
0.1009289699 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_000157_070 |
0.1019615652 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 17 |
Hb_002995_040 |
0.1044329509 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007441_280 |
0.1052078635 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 19 |
Hb_002596_060 |
0.1058694634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_089140_040 |
0.1075245851 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |