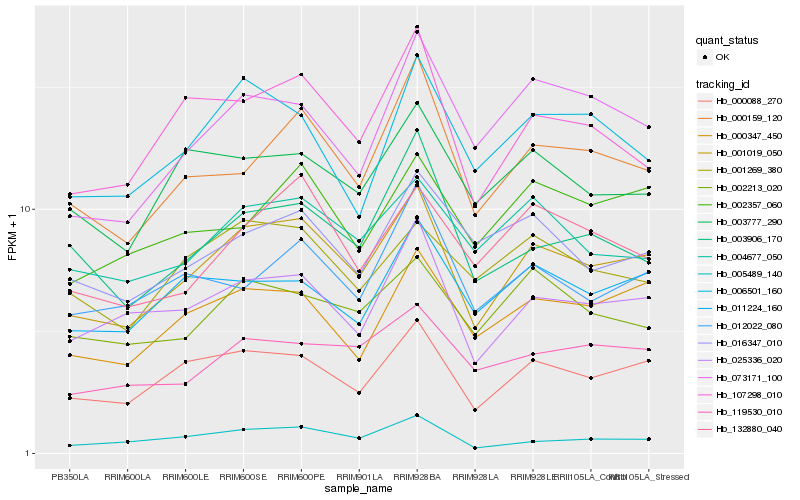
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_050 |
0.0 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 2 |
Hb_000088_270 |
0.0661382047 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 3 |
Hb_025336_020 |
0.0847554017 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 4 |
Hb_000347_450 |
0.0915844993 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000159_120 |
0.0918017797 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 6 |
Hb_107298_010 |
0.093116419 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 7 |
Hb_005489_140 |
0.0956333886 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 8 |
Hb_119530_010 |
0.0990074799 |
- |
- |
- |
| 9 |
Hb_004677_050 |
0.099420986 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_073171_100 |
0.103181975 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 11 |
Hb_011224_160 |
0.1032360419 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_002213_020 |
0.103376664 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 13 |
Hb_132880_040 |
0.1042186115 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_003777_290 |
0.1042353773 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 15 |
Hb_002357_060 |
0.1063683491 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 16 |
Hb_003906_170 |
0.1066346063 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_006501_160 |
0.1067112528 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 18 |
Hb_012022_080 |
0.107519821 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_016347_010 |
0.1075727083 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 20 |
Hb_001269_380 |
0.1078010101 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |