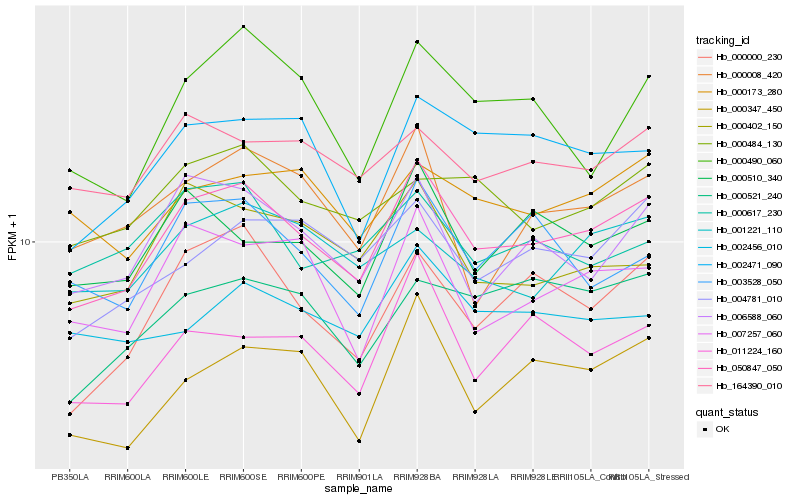
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_050847_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006588_060 |
0.0758290379 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 3 |
Hb_000000_230 |
0.0767820167 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002471_090 |
0.0818858353 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 5 |
Hb_004781_010 |
0.0827554952 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 6 |
Hb_000510_340 |
0.082798464 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 7 |
Hb_007257_060 |
0.0838913701 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 8 |
Hb_011224_160 |
0.0846309987 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 9 |
Hb_000347_450 |
0.084741579 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000521_240 |
0.0854200068 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 11 |
Hb_164390_010 |
0.0863718294 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 12 |
Hb_000484_130 |
0.0864971264 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 13 |
Hb_000008_420 |
0.0870022508 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_110 |
0.0875383833 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 15 |
Hb_003528_050 |
0.0875553079 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_000490_060 |
0.0879994157 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 17 |
Hb_000173_280 |
0.0898757005 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 18 |
Hb_000402_150 |
0.0908630331 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002456_010 |
0.0917443525 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_000617_230 |
0.0930558879 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |