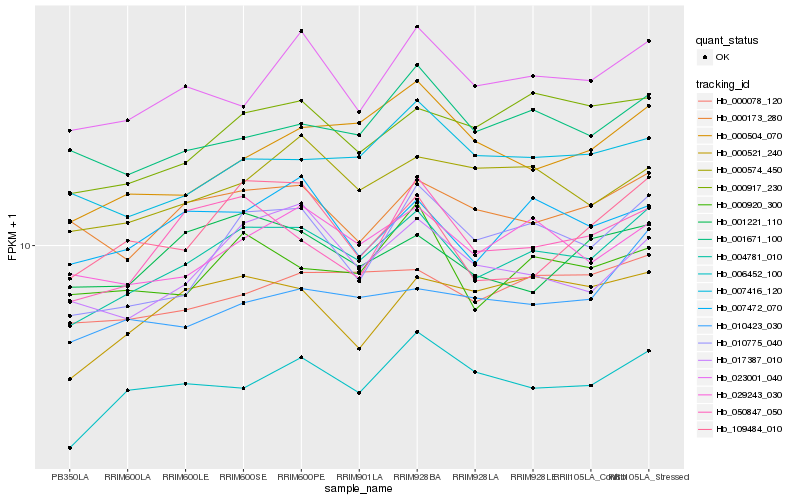
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004781_010 |
0.0 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 2 |
Hb_010775_040 |
0.0749935304 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 3 |
Hb_029243_030 |
0.0777380482 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000078_120 |
0.0800411215 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 5 |
Hb_000917_230 |
0.0807122063 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 6 |
Hb_006452_100 |
0.080896356 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 7 |
Hb_023001_040 |
0.081279656 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_017387_010 |
0.0814667773 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 9 |
Hb_050847_050 |
0.0827554952 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000920_300 |
0.083067846 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 11 |
Hb_007472_070 |
0.0851112385 |
- |
- |
cir, putative [Ricinus communis] |
| 12 |
Hb_000173_280 |
0.085837485 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 13 |
Hb_001671_100 |
0.0872600747 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 14 |
Hb_007416_120 |
0.0875895568 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 15 |
Hb_000574_450 |
0.0881110413 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 16 |
Hb_000521_240 |
0.0883228013 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 17 |
Hb_109484_010 |
0.088775967 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 18 |
Hb_000504_070 |
0.08951864 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_001221_110 |
0.089766971 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 20 |
Hb_010423_030 |
0.090131308 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |