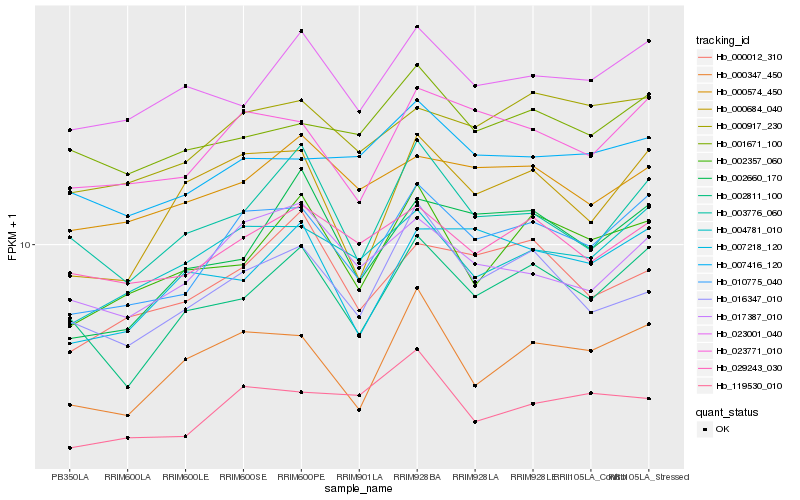
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010775_040 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 2 |
Hb_023771_010 |
0.0640977201 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 3 |
Hb_004781_010 |
0.0749935304 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 4 |
Hb_000917_230 |
0.0806623782 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 5 |
Hb_002811_100 |
0.0851450328 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 6 |
Hb_000347_450 |
0.086525147 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000684_040 |
0.0884914112 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 8 |
Hb_029243_030 |
0.0905977492 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_017387_010 |
0.0917399905 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 10 |
Hb_002357_060 |
0.0944070836 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 11 |
Hb_007416_120 |
0.0972903088 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 12 |
Hb_023001_040 |
0.0973876807 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_119530_010 |
0.098315317 |
- |
- |
- |
| 14 |
Hb_000012_310 |
0.0995096168 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001671_100 |
0.0996755595 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 16 |
Hb_003776_060 |
0.0997961886 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 17 |
Hb_000574_450 |
0.1001492842 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 18 |
Hb_002660_170 |
0.1008090562 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_007218_120 |
0.1012580038 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 20 |
Hb_016347_010 |
0.1012706112 |
- |
- |
protein transporter, putative [Ricinus communis] |