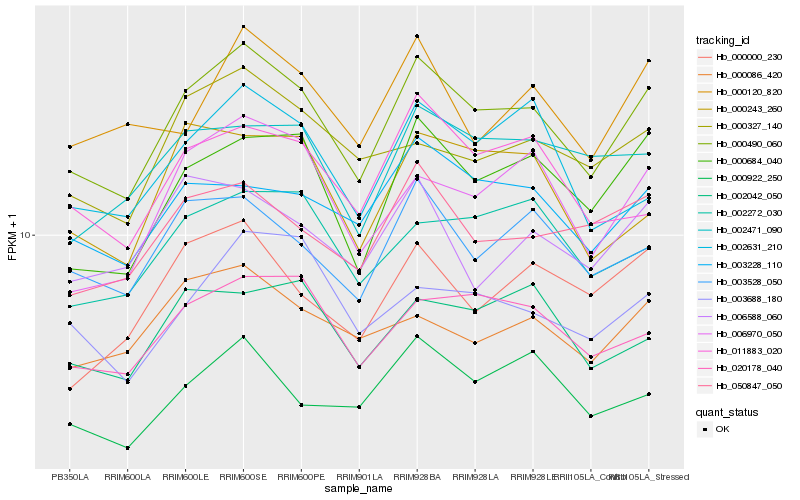
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000490_060 |
0.0 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 2 |
Hb_000684_040 |
0.07385314 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 3 |
Hb_003528_050 |
0.0826826626 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 4 |
Hb_020178_040 |
0.0829824181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006970_050 |
0.0856726307 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 6 |
Hb_003228_110 |
0.087985369 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 7 |
Hb_050847_050 |
0.0879994157 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002272_030 |
0.0935533023 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 9 |
Hb_000086_420 |
0.0945279202 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 10 |
Hb_011883_020 |
0.0945991681 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 11 |
Hb_000120_820 |
0.0950582216 |
- |
- |
dihydrodipicolinate synthase, putative [Ricinus communis] |
| 12 |
Hb_002471_090 |
0.0954115038 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 13 |
Hb_000922_250 |
0.0958081832 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_002631_210 |
0.0962513492 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 15 |
Hb_000243_260 |
0.0964683586 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000327_140 |
0.0965384972 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002042_050 |
0.0969681997 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000000_230 |
0.097352786 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003688_180 |
0.097764112 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 20 |
Hb_006588_060 |
0.0986283296 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |