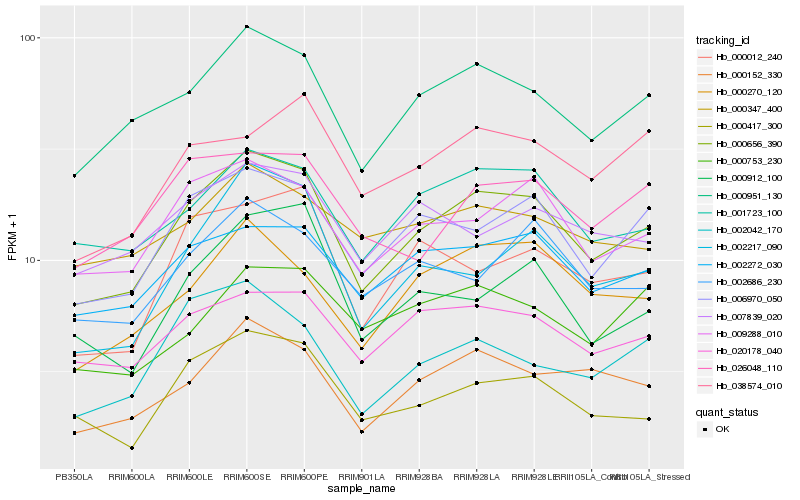
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_390 |
0.0 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000951_130 |
0.0707567297 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 3 |
Hb_006970_050 |
0.0815921182 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 4 |
Hb_009288_010 |
0.0835159125 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_001723_100 |
0.0870128491 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000152_330 |
0.0891103656 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 7 |
Hb_002272_030 |
0.0925863372 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 8 |
Hb_020178_040 |
0.09486241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000270_120 |
0.0969629548 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 10 |
Hb_000417_300 |
0.0987482961 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 11 |
Hb_002217_090 |
0.1015745666 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 12 |
Hb_002686_230 |
0.1034251515 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000012_240 |
0.1076505447 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_026048_110 |
0.107966966 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 15 |
Hb_007839_020 |
0.1102734078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000912_100 |
0.1104408334 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 17 |
Hb_002042_170 |
0.111248758 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 18 |
Hb_038574_010 |
0.1113440871 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 19 |
Hb_000753_230 |
0.1124171072 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000347_400 |
0.1126450759 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |