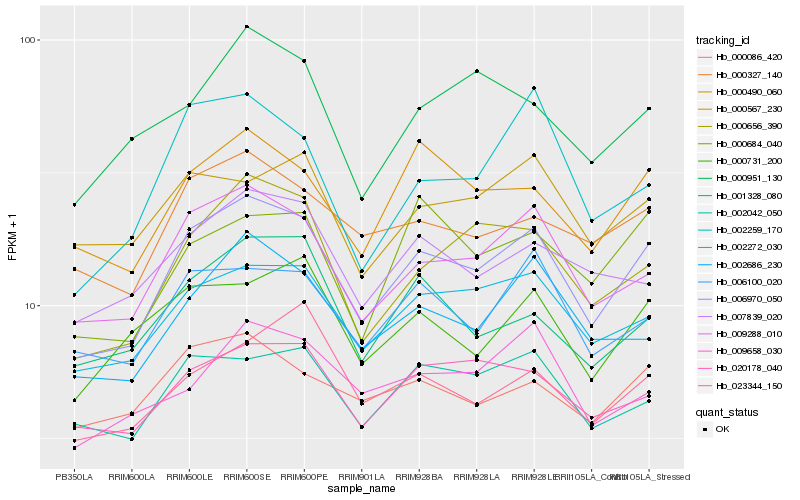
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_050 |
0.0 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 2 |
Hb_009288_010 |
0.0652770567 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_002272_030 |
0.0764380901 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 4 |
Hb_000656_390 |
0.0815921182 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000490_060 |
0.0856726307 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 6 |
Hb_002686_230 |
0.087127467 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_006100_020 |
0.0882535647 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 8 |
Hb_002042_050 |
0.0895825274 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000327_140 |
0.0906525934 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 10 |
Hb_020178_040 |
0.0914365561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000731_200 |
0.0932950562 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 12 |
Hb_007839_020 |
0.0953640702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001328_080 |
0.0963910186 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 14 |
Hb_000684_040 |
0.0973592122 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 15 |
Hb_000086_420 |
0.0986318283 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 16 |
Hb_009658_030 |
0.1005703679 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 17 |
Hb_002259_170 |
0.1005847835 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_023344_150 |
0.1013607544 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 19 |
Hb_000567_230 |
0.1013969562 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 20 |
Hb_000951_130 |
0.1014647467 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |