| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
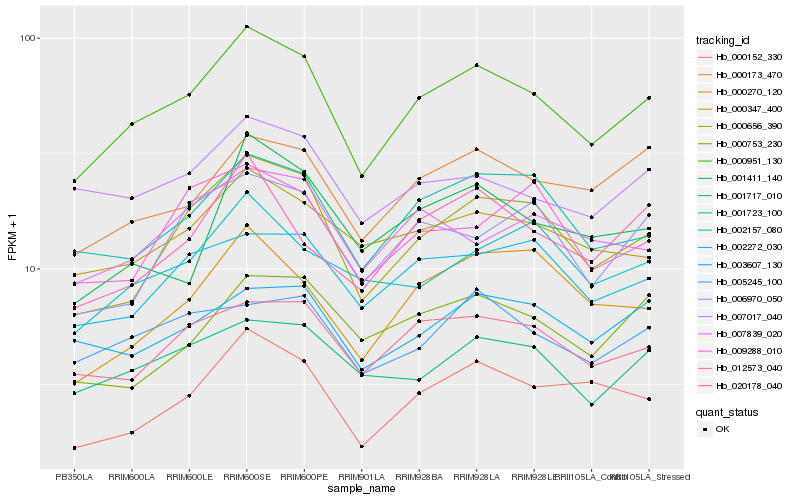
Hb_000951_130 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 2 |
Hb_000656_390 |
0.0707567297 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001723_100 |
0.0800669995 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 4 |
Hb_020178_040 |
0.0835642976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000173_470 |
0.0889743882 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_000347_400 |
0.0899581788 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002272_030 |
0.0909929044 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 8 |
Hb_001717_010 |
0.0910062878 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 9 |
Hb_000152_330 |
0.0926120071 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 10 |
Hb_005245_100 |
0.0955871156 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_007017_040 |
0.0982270153 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 12 |
Hb_003607_130 |
0.0990031624 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 13 |
Hb_012573_040 |
0.0995540062 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 14 |
Hb_007839_020 |
0.100081906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006970_050 |
0.1014647467 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 16 |
Hb_000270_120 |
0.1018228005 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 17 |
Hb_002157_080 |
0.1042365881 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 18 |
Hb_009288_010 |
0.1047872907 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001411_140 |
0.1051762955 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 20 |
Hb_000753_230 |
0.105380772 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |