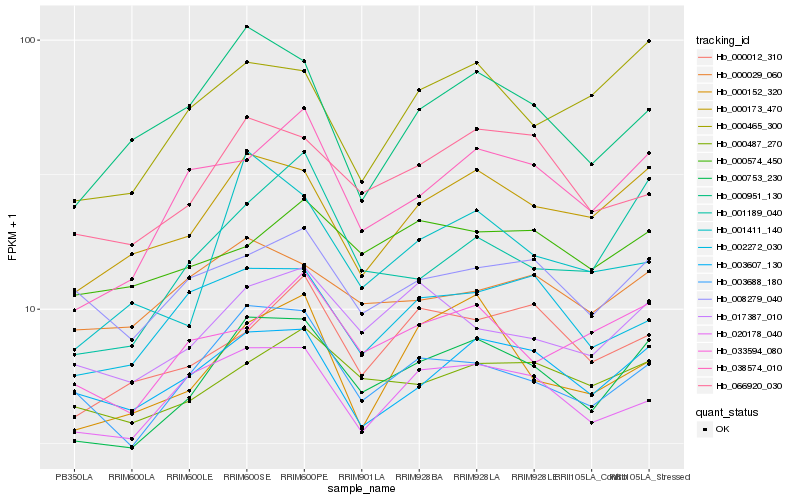
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_230 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000173_470 |
0.0764286709 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_003688_180 |
0.0846825503 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 4 |
Hb_066920_030 |
0.0888406256 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 5 |
Hb_038574_010 |
0.0900681154 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 6 |
Hb_017387_010 |
0.0907952835 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 7 |
Hb_020178_040 |
0.0967135658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003607_130 |
0.0971800818 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000487_270 |
0.1014283953 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 10 |
Hb_002272_030 |
0.1026515099 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 11 |
Hb_033594_080 |
0.1026558956 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 12 |
Hb_000465_300 |
0.1030327494 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 13 |
Hb_008279_040 |
0.104470877 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_000951_130 |
0.105380772 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 15 |
Hb_001411_140 |
0.1057830847 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 16 |
Hb_000152_320 |
0.1061201539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000012_310 |
0.1063142802 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001189_040 |
0.1069113563 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 19 |
Hb_000029_060 |
0.1087989652 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 20 |
Hb_000574_450 |
0.1101991532 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |