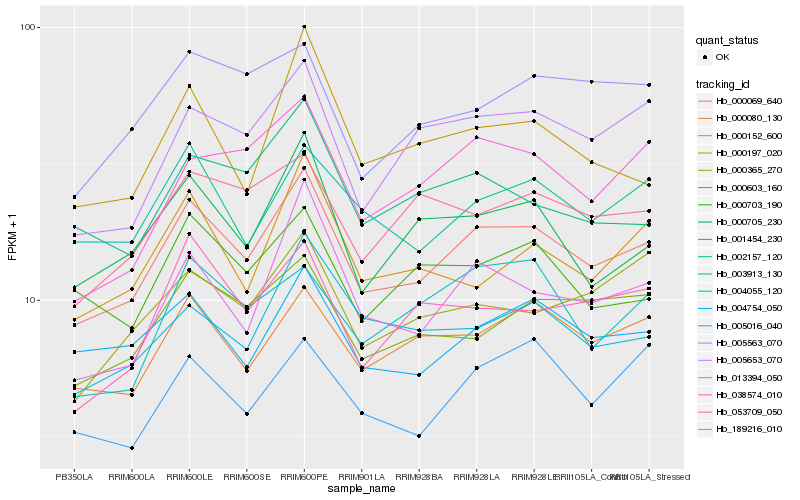
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_640 |
0.0 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 2 |
Hb_000080_130 |
0.0755837724 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 3 |
Hb_000603_160 |
0.0787465451 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 4 |
Hb_005653_070 |
0.0788718447 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 5 |
Hb_004754_050 |
0.0792449076 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 6 |
Hb_000705_230 |
0.0794312935 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 7 |
Hb_013394_050 |
0.0799750396 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 8 |
Hb_002157_120 |
0.0801317094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_189216_010 |
0.0862108222 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004055_120 |
0.086628629 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 11 |
Hb_053709_050 |
0.0872004728 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_005563_070 |
0.0890790293 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 13 |
Hb_038574_010 |
0.0894384846 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000703_190 |
0.0898105998 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000152_600 |
0.0900870906 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 16 |
Hb_001454_230 |
0.0907315243 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 17 |
Hb_000197_020 |
0.091903033 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 18 |
Hb_000365_270 |
0.0919215483 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 19 |
Hb_005016_040 |
0.0926948476 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 20 |
Hb_003913_130 |
0.0938250641 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |