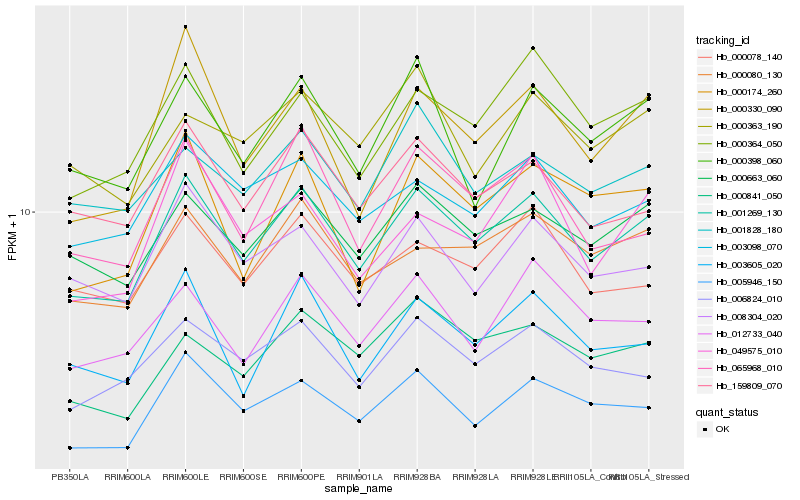
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_130 |
0.0 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 2 |
Hb_000663_060 |
0.0600866493 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 3 |
Hb_000080_130 |
0.0678781067 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 4 |
Hb_000364_050 |
0.0690997264 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003098_070 |
0.0739656215 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 6 |
Hb_006824_010 |
0.0749033702 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 7 |
Hb_005946_150 |
0.0759066094 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 8 |
Hb_159809_070 |
0.0762349539 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 9 |
Hb_065968_010 |
0.0772319322 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 10 |
Hb_008304_020 |
0.0780570361 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 11 |
Hb_012733_040 |
0.079217055 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 12 |
Hb_000330_090 |
0.0793116496 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000078_140 |
0.0793426005 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 14 |
Hb_000398_060 |
0.0802592025 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 15 |
Hb_000174_260 |
0.0807049426 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 16 |
Hb_000841_050 |
0.0816949876 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 17 |
Hb_003605_020 |
0.0828582319 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 18 |
Hb_001828_180 |
0.0838178273 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 19 |
Hb_049575_010 |
0.0862818438 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 20 |
Hb_000363_190 |
0.0868473288 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |