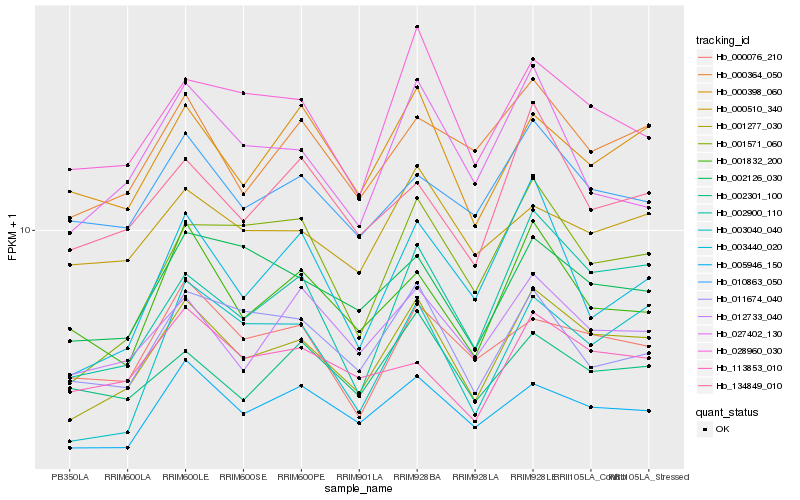
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_030 |
0.0 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Nelumbo nucifera] |
| 2 |
Hb_002900_110 |
0.0924040289 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_005946_150 |
0.0955731062 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 4 |
Hb_028960_030 |
0.1048229067 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 5 |
Hb_001571_060 |
0.1057466107 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 6 |
Hb_012733_040 |
0.1073325242 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 7 |
Hb_000364_050 |
0.1096569377 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002126_030 |
0.1100125237 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 9 |
Hb_000398_060 |
0.1107405424 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 10 |
Hb_011674_040 |
0.1125828647 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001832_200 |
0.1138225859 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_134849_010 |
0.1171269883 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 13 |
Hb_000076_210 |
0.1186715949 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_010863_050 |
0.1189371441 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 15 |
Hb_003040_040 |
0.1189565226 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002301_100 |
0.1198872341 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 17 |
Hb_003440_020 |
0.1205499882 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_027402_130 |
0.1211580974 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000510_340 |
0.1217736414 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 20 |
Hb_113853_010 |
0.122124379 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |