| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
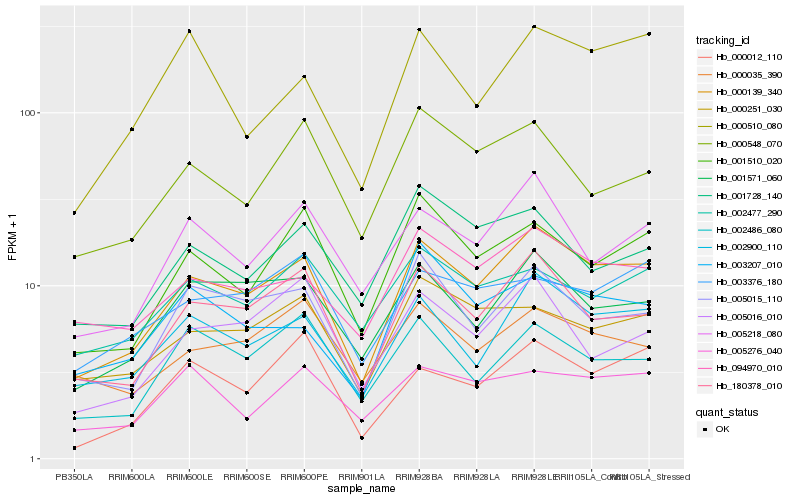
| 1 |
Hb_000139_340 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002900_110 |
0.0856994667 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_180378_010 |
0.0907678614 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 4 |
Hb_094970_010 |
0.1054939724 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005015_110 |
0.1093872676 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 6 |
Hb_001571_060 |
0.1106515456 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 7 |
Hb_005218_080 |
0.1133564848 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001510_020 |
0.1170962342 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 9 |
Hb_000251_030 |
0.1231613337 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000012_110 |
0.1284214468 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 11 |
Hb_000548_070 |
0.1317273303 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 12 |
Hb_000510_080 |
0.1321980098 |
- |
- |
hypothetical protein CICLE_v10021453mg [Citrus clementina] |
| 13 |
Hb_001728_140 |
0.1329690233 |
- |
- |
- |
| 14 |
Hb_005276_040 |
0.1331177841 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 15 |
Hb_002477_290 |
0.1331453644 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000035_390 |
0.1335958618 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 17 |
Hb_005016_010 |
0.1338648753 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 18 |
Hb_003376_180 |
0.1340378083 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 19 |
Hb_002486_080 |
0.1342095176 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 20 |
Hb_003207_010 |
0.1345189089 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |