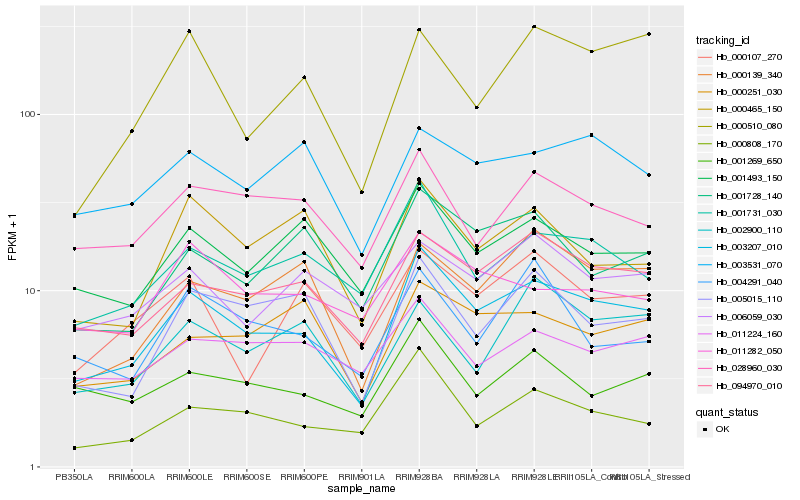
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003207_010 |
0.0 |
- |
- |
PREDICTED: phospholipase D beta 1-like [Jatropha curcas] |
| 2 |
Hb_094970_010 |
0.1058633288 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001731_030 |
0.118089534 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 4 |
Hb_005015_110 |
0.1287450034 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 5 |
Hb_000808_170 |
0.1292203874 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 6 |
Hb_001493_150 |
0.1319845174 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 7 |
Hb_000139_340 |
0.1345189089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001728_140 |
0.134879375 |
- |
- |
- |
| 9 |
Hb_028960_030 |
0.1379137197 |
- |
- |
Short-chain dehydrogenase-reductase B [Theobroma cacao] |
| 10 |
Hb_001269_650 |
0.1399172052 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_000251_030 |
0.1415071946 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000465_150 |
0.1436103519 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 13 |
Hb_003531_070 |
0.1441469376 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000510_080 |
0.1445860856 |
- |
- |
hypothetical protein CICLE_v10021453mg [Citrus clementina] |
| 15 |
Hb_011224_160 |
0.147061026 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 16 |
Hb_011282_050 |
0.148649146 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_006059_030 |
0.1511729461 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000107_270 |
0.1514907881 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_002900_110 |
0.1515177086 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_004291_040 |
0.1517979946 |
- |
- |
hexokinase [Manihot esculenta] |