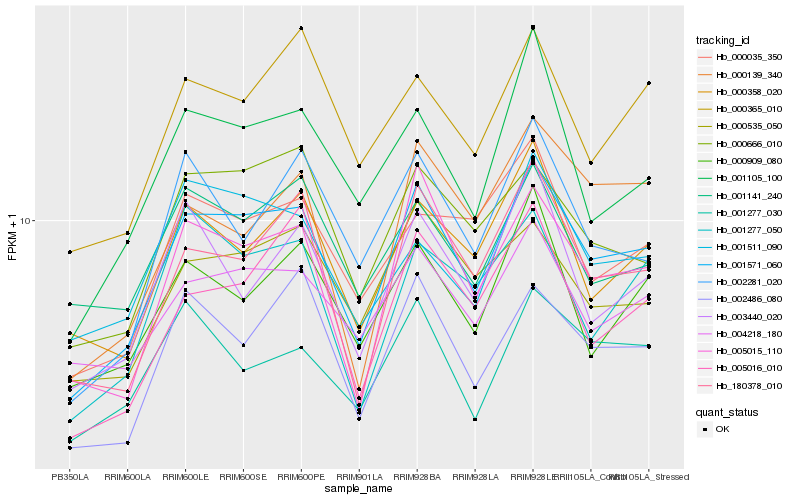
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001571_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 2 |
Hb_005015_110 |
0.0831993631 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 3 |
Hb_180378_010 |
0.0903039561 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 4 |
Hb_004218_180 |
0.0914183597 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 5 |
Hb_000666_010 |
0.0928185579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002486_080 |
0.0931418635 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 7 |
Hb_000535_050 |
0.095481974 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 8 |
Hb_000358_020 |
0.0977799527 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 9 |
Hb_001105_100 |
0.0987302035 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 10 |
Hb_000365_010 |
0.0997145814 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 11 |
Hb_000909_080 |
0.1015145562 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 12 |
Hb_000035_350 |
0.1028440951 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001277_050 |
0.1049121642 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001511_090 |
0.1054632471 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 15 |
Hb_001277_030 |
0.1057466107 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Nelumbo nucifera] |
| 16 |
Hb_003440_020 |
0.1074297058 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_005016_010 |
0.1079562514 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 18 |
Hb_001141_240 |
0.1082437993 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002281_020 |
0.1097642229 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 20 |
Hb_000139_340 |
0.1106515456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |