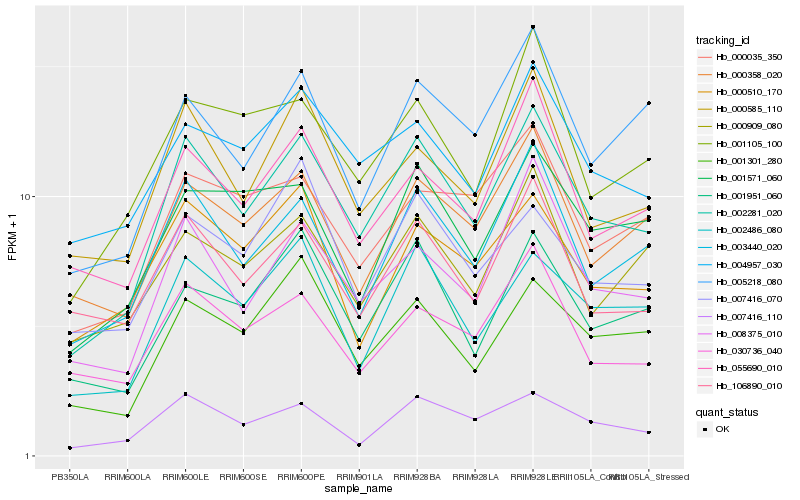
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002281_020 |
0.0 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 2 |
Hb_003440_020 |
0.0840544291 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_008375_010 |
0.0923857149 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 4 |
Hb_002486_080 |
0.0925976923 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 5 |
Hb_007416_110 |
0.0949650044 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000585_110 |
0.1001296057 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 7 |
Hb_001301_280 |
0.1014807273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001951_060 |
0.1019786508 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 9 |
Hb_005218_080 |
0.1022677037 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001105_100 |
0.1038955822 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 11 |
Hb_055690_010 |
0.1052176043 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004957_030 |
0.105379324 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 13 |
Hb_000358_020 |
0.1059122835 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 14 |
Hb_001571_060 |
0.1097642229 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 15 |
Hb_106890_010 |
0.1118746875 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_030736_040 |
0.112508766 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000035_350 |
0.1130380553 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000909_080 |
0.1131695874 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 19 |
Hb_007416_070 |
0.1163419383 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 20 |
Hb_000510_170 |
0.1171058591 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |