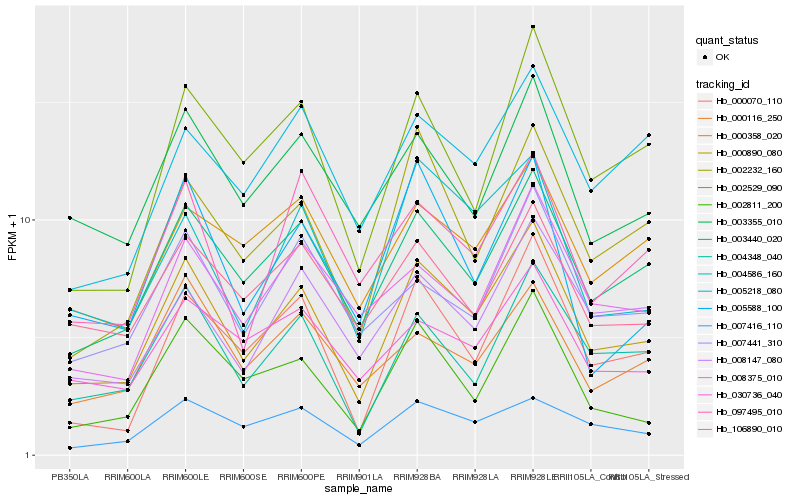
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_080 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 2 |
Hb_003440_020 |
0.0835654071 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_004348_040 |
0.1008415288 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 4 |
Hb_002232_160 |
0.1079806587 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 5 |
Hb_000070_110 |
0.1109543982 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002529_090 |
0.1122735659 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_007416_110 |
0.1128768118 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004586_160 |
0.1143853297 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003355_010 |
0.1161917236 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 10 |
Hb_002811_200 |
0.1169210386 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 11 |
Hb_106890_010 |
0.1197619219 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_030736_040 |
0.1201976191 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008147_080 |
0.1204452403 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000358_020 |
0.1214061009 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 15 |
Hb_008375_010 |
0.123686022 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 16 |
Hb_007441_310 |
0.1240013165 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005218_080 |
0.1264808243 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000116_250 |
0.1273318941 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 19 |
Hb_097495_010 |
0.1283889606 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005588_100 |
0.1289886177 |
- |
- |
protein phosphatase, putative [Ricinus communis] |