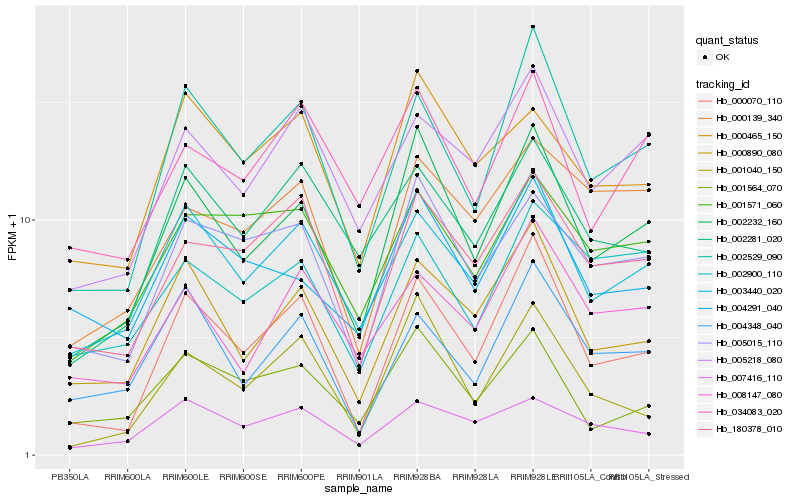
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_160 |
0.0 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 2 |
Hb_000890_080 |
0.1079806587 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 3 |
Hb_000070_110 |
0.1085462799 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003440_020 |
0.1086011718 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002529_090 |
0.1096965779 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_005015_110 |
0.1111242394 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 7 |
Hb_001564_070 |
0.1220107306 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 8 |
Hb_001040_150 |
0.127136396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004291_040 |
0.127839943 |
- |
- |
hexokinase [Manihot esculenta] |
| 10 |
Hb_180378_010 |
0.1279257848 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 11 |
Hb_000465_150 |
0.1322069816 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 12 |
Hb_005218_080 |
0.1326328844 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_034083_020 |
0.1353338948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007416_110 |
0.1384346232 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002281_020 |
0.1396557115 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 16 |
Hb_004348_040 |
0.1418124655 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 17 |
Hb_001571_060 |
0.1418744228 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 18 |
Hb_002900_110 |
0.1430695548 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_000139_340 |
0.1431824114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008147_080 |
0.1433415513 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |