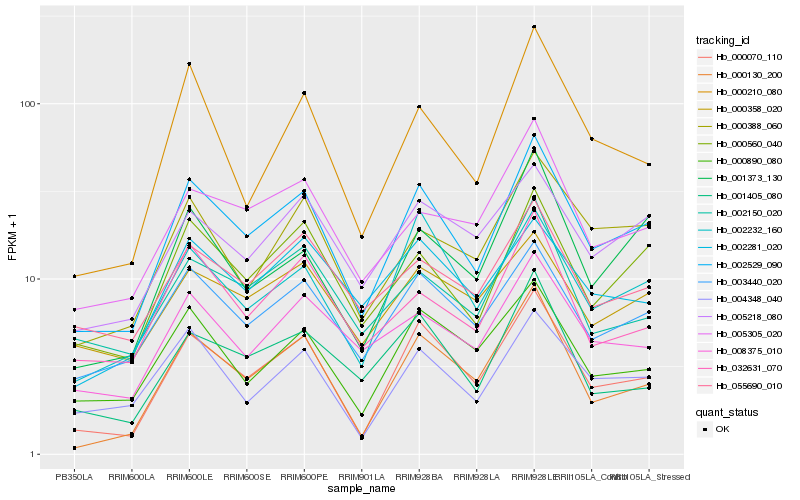
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002529_090 |
0.0 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_000070_110 |
0.0772338003 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000560_040 |
0.0881898332 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003440_020 |
0.089152784 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_004348_040 |
0.1067511233 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 6 |
Hb_000130_200 |
0.1080503773 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 7 |
Hb_002232_160 |
0.1096965779 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 8 |
Hb_000388_060 |
0.1114376157 |
- |
- |
fructokinase [Manihot esculenta] |
| 9 |
Hb_000890_080 |
0.1122735659 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 10 |
Hb_008375_010 |
0.112479968 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 11 |
Hb_005218_080 |
0.1137204687 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_005305_020 |
0.1139137771 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001373_130 |
0.1184104457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002150_020 |
0.1188882481 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_032631_070 |
0.1192747369 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_001405_080 |
0.1197447467 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 17 |
Hb_055690_010 |
0.1201899193 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000358_020 |
0.1212590534 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 19 |
Hb_000210_080 |
0.1219289974 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002281_020 |
0.1221002033 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |