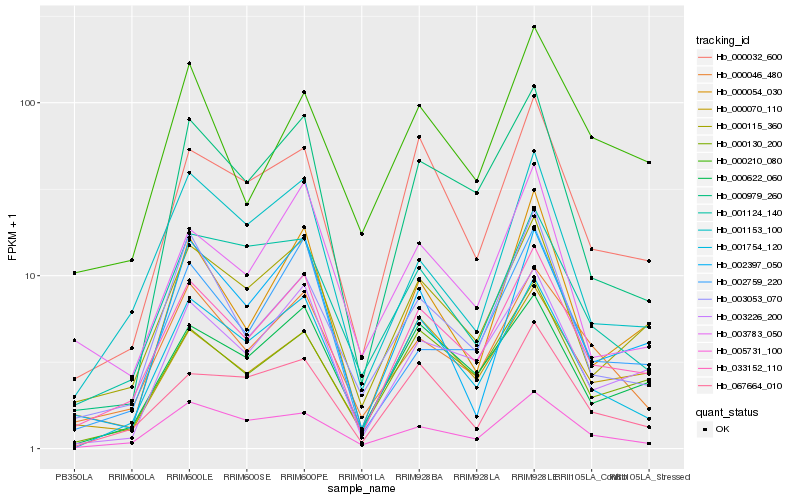
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033152_110 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 2 |
Hb_000115_360 |
0.0859902406 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 3 |
Hb_003226_200 |
0.0970537434 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 4 |
Hb_000622_060 |
0.1007210949 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000130_200 |
0.1010426222 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 6 |
Hb_001754_120 |
0.1030046197 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 7 |
Hb_000046_480 |
0.1032597777 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 8 |
Hb_000032_600 |
0.1095278879 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 9 |
Hb_005731_100 |
0.1177273241 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 10 |
Hb_000070_110 |
0.1221346793 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000054_030 |
0.1238332296 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 12 |
Hb_067664_010 |
0.1250710686 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 13 |
Hb_000979_260 |
0.1264955753 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 14 |
Hb_002759_220 |
0.1287716155 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 15 |
Hb_001153_100 |
0.1306629435 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 16 |
Hb_003053_070 |
0.1346701498 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001124_140 |
0.1353354193 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 18 |
Hb_003783_050 |
0.1358873956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000210_080 |
0.1385277753 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002397_050 |
0.1393998049 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |