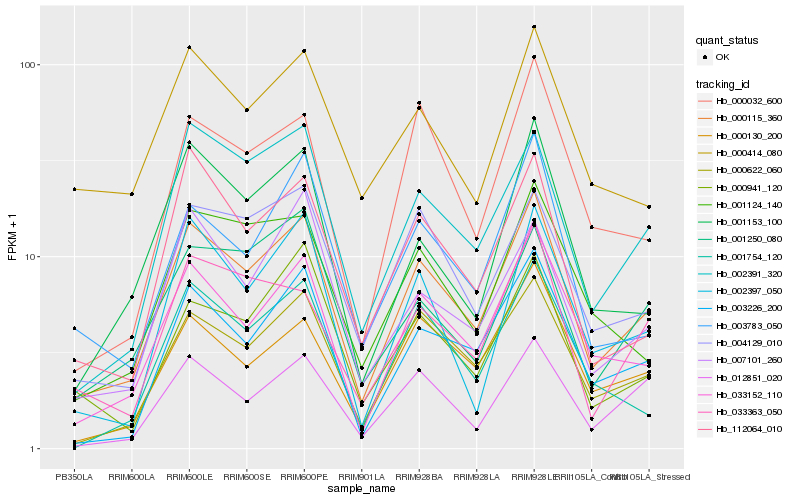
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_360 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 2 |
Hb_033152_110 |
0.0859902406 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 3 |
Hb_000622_060 |
0.0936318372 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002391_320 |
0.1037618989 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 5 |
Hb_003226_200 |
0.1066349367 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 6 |
Hb_004129_010 |
0.1169688306 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_112064_010 |
0.119836662 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 8 |
Hb_000130_200 |
0.1206358257 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 9 |
Hb_001153_100 |
0.1255529754 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 10 |
Hb_000941_120 |
0.1260684628 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 11 |
Hb_007101_260 |
0.1266656837 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 12 |
Hb_012851_020 |
0.1274681434 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000032_600 |
0.1275770319 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 14 |
Hb_001124_140 |
0.1276794597 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 15 |
Hb_002397_050 |
0.12778712 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001250_080 |
0.1293089575 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 17 |
Hb_003783_050 |
0.1300164091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001754_120 |
0.1304849507 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 19 |
Hb_000414_080 |
0.1317034198 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_033363_050 |
0.1324560501 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 isoform X2 [Jatropha curcas] |