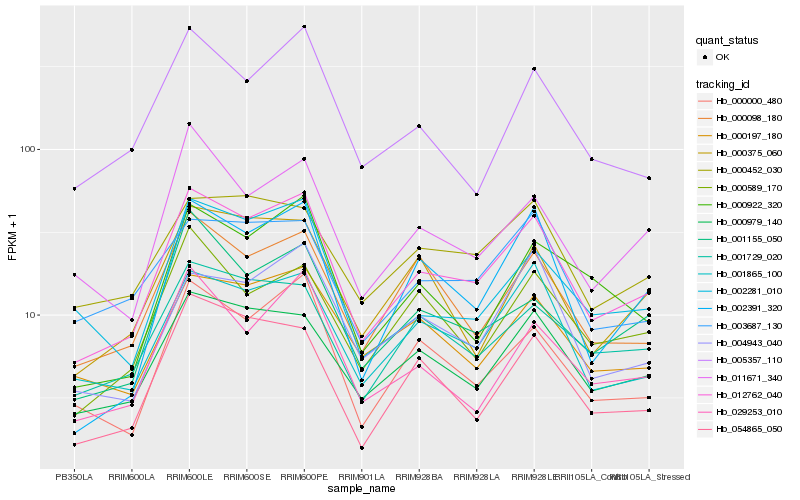
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012762_040 |
0.0 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 2 |
Hb_000979_140 |
0.0928915058 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 3 |
Hb_001865_100 |
0.099098984 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 4 |
Hb_000000_480 |
0.1017543303 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001729_020 |
0.1063942718 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000197_180 |
0.1083499926 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 7 |
Hb_000098_180 |
0.1097481074 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002391_320 |
0.1100251917 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 9 |
Hb_002281_010 |
0.1141710428 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 10 |
Hb_000922_320 |
0.1159753887 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 11 |
Hb_005357_110 |
0.116240929 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 12 |
Hb_054865_050 |
0.1171716773 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 13 |
Hb_004943_040 |
0.1176302913 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011671_340 |
0.1185418731 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 15 |
Hb_003687_130 |
0.1190677815 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 16 |
Hb_000589_170 |
0.1193331121 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000375_060 |
0.1202315784 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 18 |
Hb_001155_050 |
0.1202443481 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 19 |
Hb_000452_030 |
0.120821181 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_029253_010 |
0.1210256019 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |