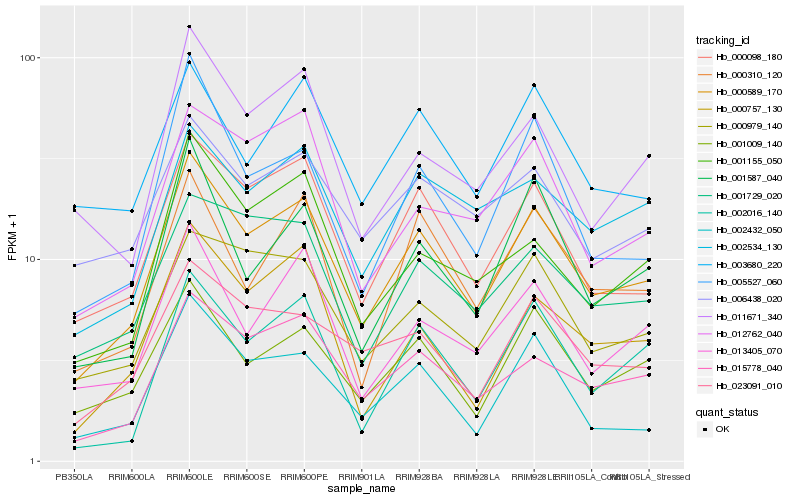
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000589_170 |
0.0 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000098_180 |
0.084584844 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 3 |
Hb_015778_040 |
0.0885676387 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 4 |
Hb_001009_140 |
0.0964193668 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 5 |
Hb_023091_010 |
0.1078784405 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 6 |
Hb_013405_070 |
0.1084145606 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 7 |
Hb_000979_140 |
0.1100429025 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 8 |
Hb_001587_040 |
0.1120228804 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001729_020 |
0.1130280967 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001155_050 |
0.1139172936 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 11 |
Hb_000310_120 |
0.1141868111 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 12 |
Hb_002534_130 |
0.1141888157 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 13 |
Hb_005527_060 |
0.1148889636 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_002432_050 |
0.1164539033 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 15 |
Hb_000757_130 |
0.1168690332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011671_340 |
0.1184099368 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 17 |
Hb_002016_140 |
0.1184321099 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 18 |
Hb_006438_020 |
0.1187276526 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 19 |
Hb_012762_040 |
0.1193331121 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 20 |
Hb_003680_220 |
0.1197324822 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |