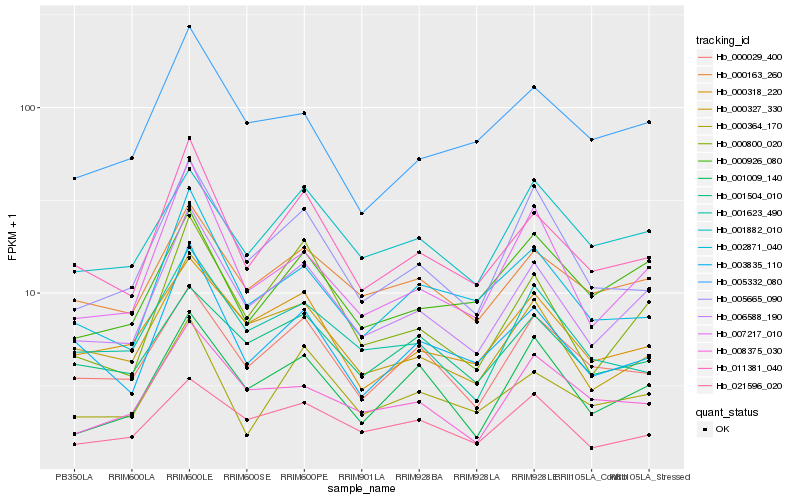
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_190 |
0.0 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 2 |
Hb_011381_040 |
0.0758720921 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000327_330 |
0.0847606327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000800_020 |
0.086433143 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 5 |
Hb_007217_010 |
0.0865753478 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000163_260 |
0.0910028 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 7 |
Hb_001009_140 |
0.0990613081 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 8 |
Hb_000318_220 |
0.0994071263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005665_090 |
0.1007396363 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 10 |
Hb_000029_400 |
0.1029418156 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002871_040 |
0.1034213279 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 12 |
Hb_003835_110 |
0.1051287808 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_021596_020 |
0.1056843287 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 14 |
Hb_008375_030 |
0.1071148778 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 15 |
Hb_001504_010 |
0.1087643354 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 16 |
Hb_005332_080 |
0.1098487137 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000926_080 |
0.1103695702 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 18 |
Hb_001882_010 |
0.1109625213 |
- |
- |
- |
| 19 |
Hb_001623_490 |
0.1111488438 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000364_170 |
0.1128679387 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |