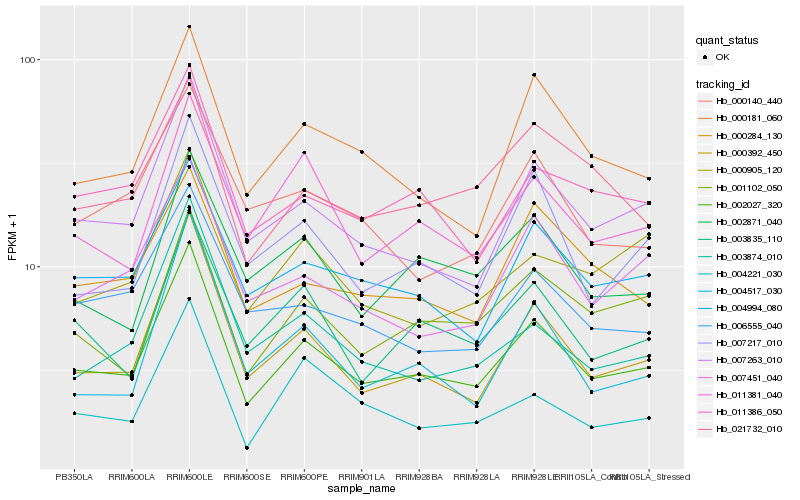
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_320 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 2 |
Hb_007263_010 |
0.0744938604 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 3 |
Hb_000392_450 |
0.0805999333 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004994_080 |
0.0897696744 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_011386_050 |
0.0961181635 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_011381_040 |
0.1000485584 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000181_060 |
0.1001417762 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 8 |
Hb_007217_010 |
0.1011273047 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003835_110 |
0.1016169398 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001102_050 |
0.1047330685 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 11 |
Hb_007451_040 |
0.1056880956 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 12 |
Hb_006555_040 |
0.107333526 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 13 |
Hb_002871_040 |
0.1085382346 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 14 |
Hb_000905_120 |
0.1127462518 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 15 |
Hb_003874_010 |
0.113008155 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000284_130 |
0.1154863474 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 17 |
Hb_004517_030 |
0.1189729238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004221_030 |
0.1191862124 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000140_440 |
0.1195917361 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 20 |
Hb_021732_010 |
0.1197050372 |
- |
- |
biotin carboxylase [Vernicia fordii] |