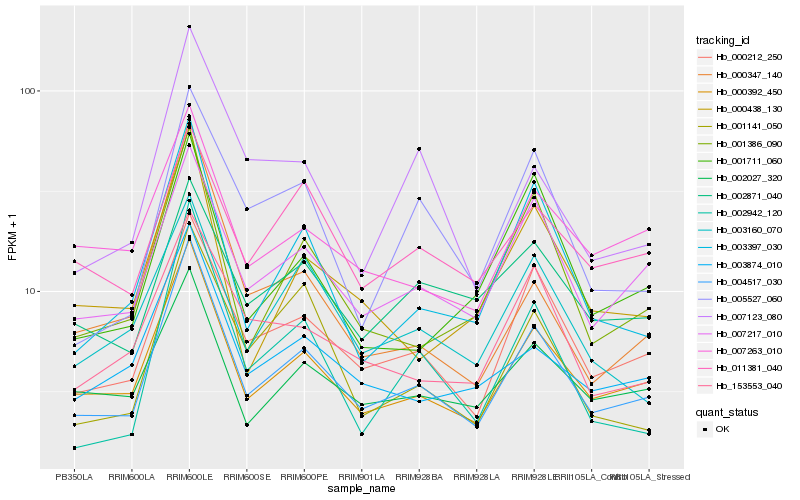
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004517_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000392_450 |
0.0617887328 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000212_250 |
0.0876901461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007217_010 |
0.1045802418 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001386_090 |
0.1094271054 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 6 |
Hb_003397_030 |
0.1107583122 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003874_010 |
0.11613058 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002027_320 |
0.1189729238 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 9 |
Hb_153553_040 |
0.1268691597 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 10 |
Hb_000438_130 |
0.1280091487 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 11 |
Hb_003160_070 |
0.1305552979 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 12 |
Hb_007263_010 |
0.1323541655 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_005527_060 |
0.1359504736 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_002942_120 |
0.136690319 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 15 |
Hb_007123_080 |
0.1377503295 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 16 |
Hb_011381_040 |
0.1397756206 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002871_040 |
0.1397855235 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 18 |
Hb_000347_140 |
0.1402047243 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_001141_050 |
0.1409363378 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001711_060 |
0.1431366591 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |