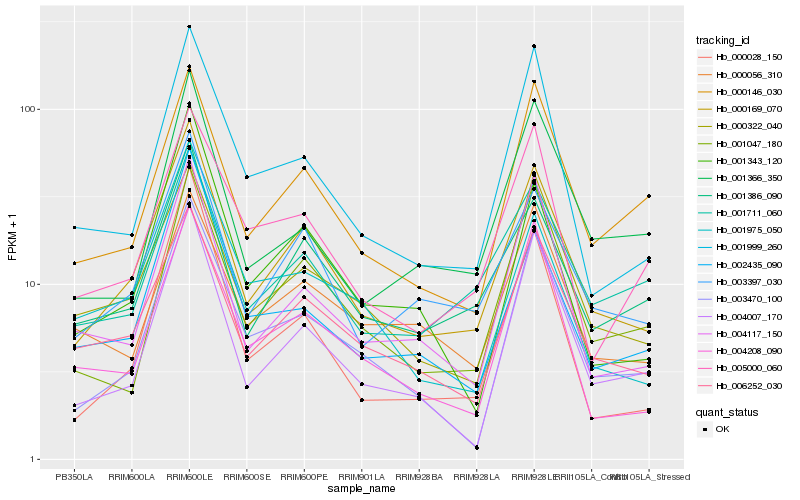
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000169_070 |
0.0 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 2 |
Hb_003397_030 |
0.081803629 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001386_090 |
0.0998574247 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 4 |
Hb_006252_030 |
0.0999476439 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 5 |
Hb_000322_040 |
0.1166564203 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003470_100 |
0.1207039405 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_004007_170 |
0.1230900762 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_000146_030 |
0.1257870087 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001711_060 |
0.1263767079 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 10 |
Hb_004117_150 |
0.1275354109 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 11 |
Hb_001999_260 |
0.1283899161 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001047_180 |
0.1290883269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001343_120 |
0.1305613896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004208_090 |
0.1312782399 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 15 |
Hb_002435_090 |
0.133604314 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005000_060 |
0.1355043156 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 17 |
Hb_001975_050 |
0.1370068712 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 18 |
Hb_000028_150 |
0.1407670554 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 19 |
Hb_001366_350 |
0.1408500075 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 20 |
Hb_000056_310 |
0.1417812511 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |