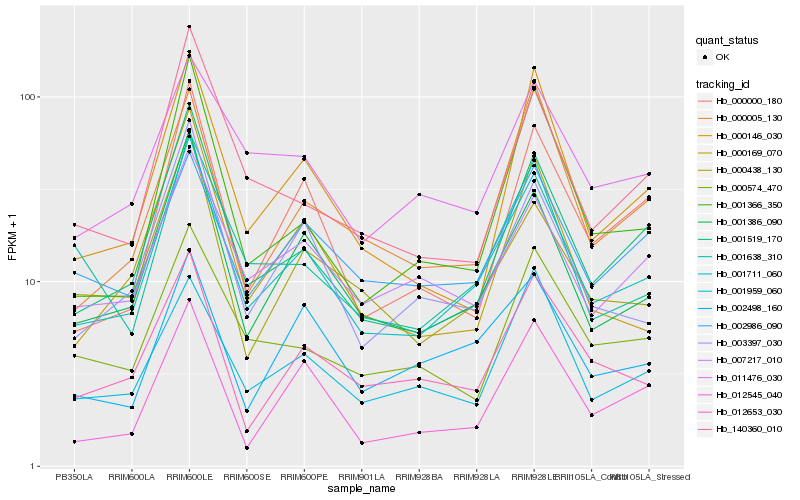
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001711_060 |
0.0 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 2 |
Hb_001386_090 |
0.0813222243 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 3 |
Hb_001519_170 |
0.0852856357 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003397_030 |
0.1105948908 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_012653_030 |
0.1184791258 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000146_030 |
0.1190232843 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000438_130 |
0.1194126823 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 8 |
Hb_007217_010 |
0.1203224424 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001366_350 |
0.1210602957 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 10 |
Hb_001959_060 |
0.1221747988 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_012545_040 |
0.1250398587 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_011476_030 |
0.1256105018 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 13 |
Hb_000169_070 |
0.1263767079 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 14 |
Hb_001638_310 |
0.1270028657 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 15 |
Hb_000000_180 |
0.1274831128 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_000574_470 |
0.1288013723 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_002498_160 |
0.1289152224 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 18 |
Hb_140360_010 |
0.129078383 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000005_130 |
0.1312450239 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 20 |
Hb_002986_090 |
0.1322762721 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |