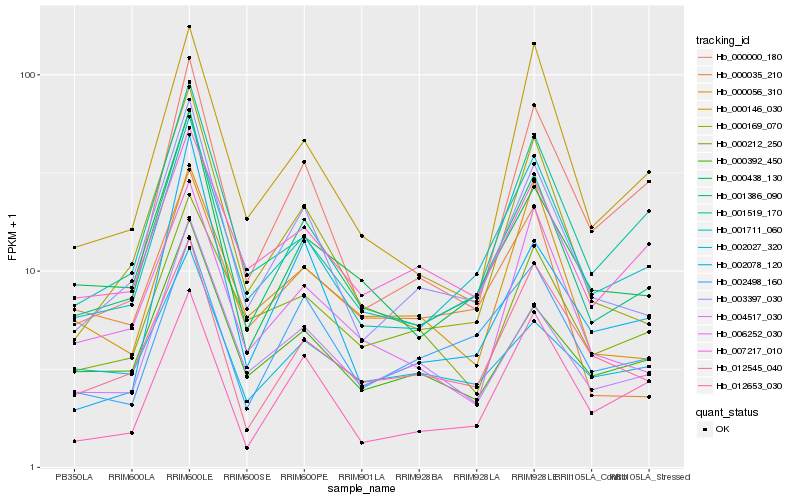
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001386_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 2 |
Hb_003397_030 |
0.0764776702 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000438_130 |
0.0779877746 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 4 |
Hb_001711_060 |
0.0813222243 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 5 |
Hb_000169_070 |
0.0998574247 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 6 |
Hb_004517_030 |
0.1094271054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000392_450 |
0.114708194 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001519_170 |
0.1178998811 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_006252_030 |
0.1258555171 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 10 |
Hb_012653_030 |
0.128551134 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_007217_010 |
0.1316598589 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000212_250 |
0.1383140928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000000_180 |
0.1389772526 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_000146_030 |
0.1399260625 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 15 |
Hb_012545_040 |
0.140219008 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_002498_160 |
0.1421985525 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 17 |
Hb_002078_120 |
0.1426864454 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 18 |
Hb_000056_310 |
0.1439425628 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 19 |
Hb_002027_320 |
0.1455226777 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 20 |
Hb_000035_210 |
0.1455434153 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |