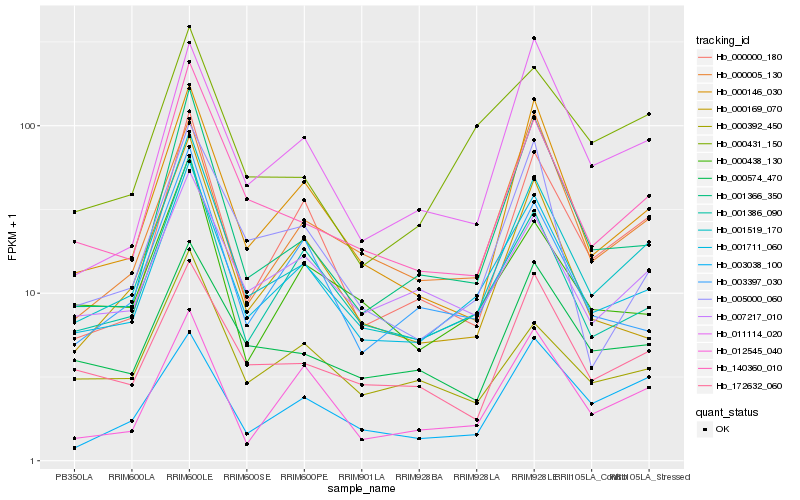
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001519_170 |
0.0 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001711_060 |
0.0852856357 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 3 |
Hb_140360_010 |
0.0867850568 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_000146_030 |
0.1099367685 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000000_180 |
0.11143829 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_001366_350 |
0.1144912271 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 7 |
Hb_001386_090 |
0.1178998811 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 8 |
Hb_000574_470 |
0.1274898248 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_172632_060 |
0.1288955862 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003038_100 |
0.1398082735 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_012545_040 |
0.1418067467 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000431_150 |
0.1422416257 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 13 |
Hb_007217_010 |
0.1427367774 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000392_450 |
0.1433831148 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000438_130 |
0.1438325912 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 16 |
Hb_000169_070 |
0.1447524745 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 17 |
Hb_000005_130 |
0.1451092089 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 18 |
Hb_011114_020 |
0.1457174712 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_003397_030 |
0.1463651292 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005000_060 |
0.1467182134 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |