| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
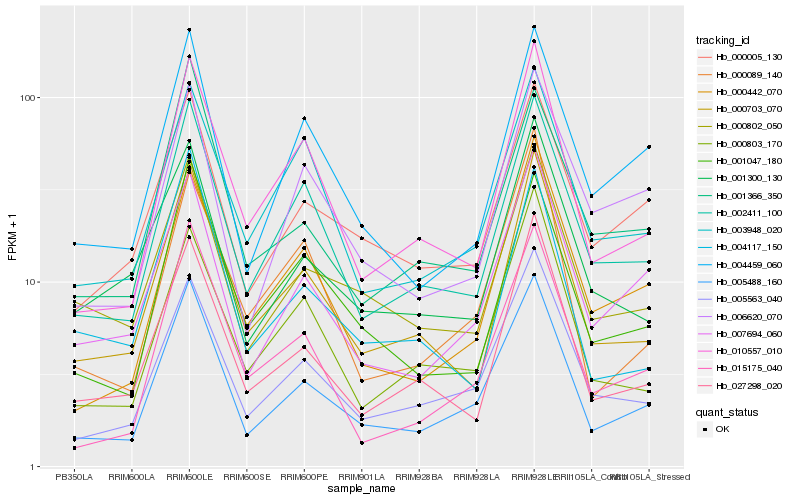
Hb_005488_160 |
0.0 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_005563_040 |
0.0917862433 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002411_100 |
0.1020736786 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 4 |
Hb_007694_060 |
0.1053195407 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 5 |
Hb_015175_040 |
0.1076660992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010557_010 |
0.1087413713 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000703_070 |
0.112466896 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004459_060 |
0.1152255584 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_001047_180 |
0.1165880576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000005_130 |
0.1170789283 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 11 |
Hb_027298_020 |
0.117814389 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 12 |
Hb_000442_070 |
0.119645223 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 13 |
Hb_000802_050 |
0.1199073188 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 14 |
Hb_001366_350 |
0.1235657538 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 15 |
Hb_000803_170 |
0.127536412 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 16 |
Hb_006620_070 |
0.1307538618 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 17 |
Hb_003948_020 |
0.1307971388 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 18 |
Hb_001300_130 |
0.1332789058 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 19 |
Hb_000089_140 |
0.1374929022 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004117_150 |
0.138025559 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |