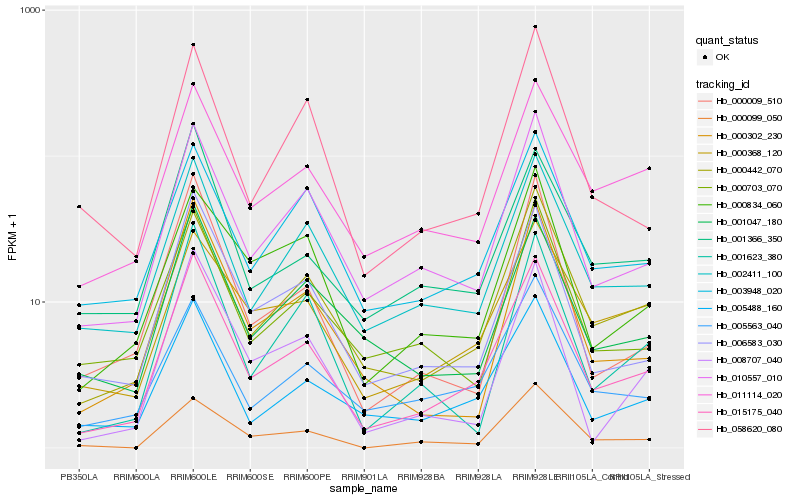
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015175_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006583_030 |
0.106409799 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 3 |
Hb_005488_160 |
0.1076660992 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_010557_010 |
0.1094078096 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000442_070 |
0.1100717898 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 6 |
Hb_002411_100 |
0.1194007857 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 7 |
Hb_000302_230 |
0.121227905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000009_510 |
0.1266913259 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 9 |
Hb_001366_350 |
0.1296829402 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 10 |
Hb_001047_180 |
0.1299605694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001623_380 |
0.1336289858 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_011114_020 |
0.1348440443 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_005563_040 |
0.1354622163 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000703_070 |
0.1389244183 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008707_040 |
0.1391098572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_058620_080 |
0.141591472 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000099_050 |
0.141820235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003948_020 |
0.1434487132 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 19 |
Hb_000834_060 |
0.1439044671 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000368_120 |
0.1465157491 |
- |
- |
PREDICTED: uncharacterized protein LOC105633862 [Jatropha curcas] |