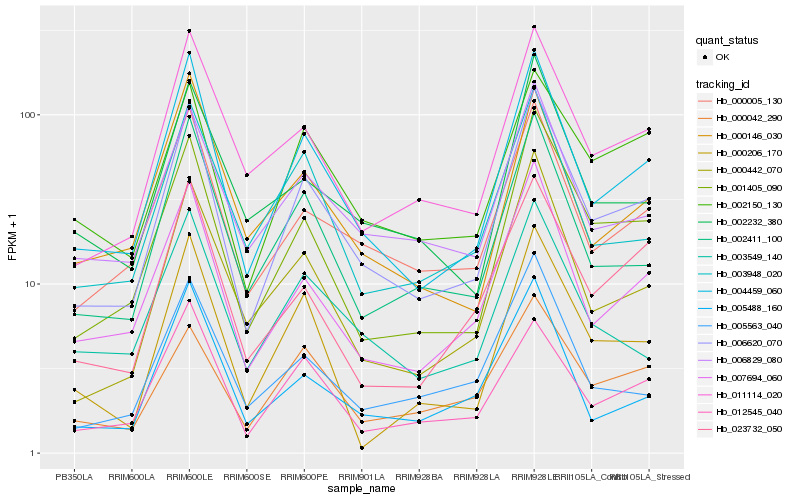
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006620_070 |
0.0 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 2 |
Hb_004459_060 |
0.0555763405 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001405_090 |
0.0870044491 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007694_060 |
0.0979449137 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000005_130 |
0.0985297573 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 6 |
Hb_002411_100 |
0.1039158969 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 7 |
Hb_000442_070 |
0.1048778517 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 8 |
Hb_011114_020 |
0.1061720611 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_012545_040 |
0.1116311018 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_006829_080 |
0.1180112952 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 11 |
Hb_002150_130 |
0.1209431124 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_003948_020 |
0.1227286419 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 13 |
Hb_003549_140 |
0.125039405 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000042_290 |
0.1252493458 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 15 |
Hb_023732_050 |
0.1260668 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_005563_040 |
0.1292317791 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002232_380 |
0.1295952282 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_005488_160 |
0.1307538618 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000146_030 |
0.1307753553 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000206_170 |
0.1310260441 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |